Imaging mass cytometry analysis of Becker muscular dystrophy muscle samples reveals different stages of muscle degeneration
- PMID: 38336890
- PMCID: PMC10858026
- DOI: 10.1038/s41598-024-51906-x
Imaging mass cytometry analysis of Becker muscular dystrophy muscle samples reveals different stages of muscle degeneration
Abstract
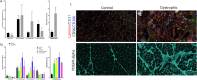
Becker muscular dystrophy (BMD) is characterised by fiber loss and expansion of fibrotic and adipose tissue. Several cells interact locally in what is known as the degenerative niche. We analysed muscle biopsies of controls and BMD patients at early, moderate and advanced stages of progression using Hyperion imaging mass cytometry (IMC) by labelling single sections with 17 markers identifying different components of the muscle. We developed a software for analysing IMC images and studied changes in the muscle composition and spatial correlations between markers across disease progression. We found a strong correlation between collagen-I and the area of stroma, collagen-VI, adipose tissue, and M2-macrophages number. There was a negative correlation between the area of collagen-I and the number of satellite cells (SCs), fibres and blood vessels. The comparison between fibrotic and non-fibrotic areas allowed to study the disease process in detail. We found structural differences among non-fibrotic areas from control and patients, being these latter characterized by increase in CTGF and in M2-macrophages and decrease in fibers and blood vessels. IMC enables to study of changes in tissue structure along disease progression, spatio-temporal correlations and opening the door to better understand new potential pathogenic pathways in human samples.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

