Mitochondrial outer membrane integrity regulates a ubiquitin-dependent and NF-κB-mediated inflammatory response
- PMID: 38337057
- PMCID: PMC10943237
- DOI: 10.1038/s44318-024-00044-1
Mitochondrial outer membrane integrity regulates a ubiquitin-dependent and NF-κB-mediated inflammatory response
Abstract
Mitochondrial outer membrane permeabilisation (MOMP) is often essential for apoptosis, by enabling cytochrome c release that leads to caspase activation and rapid cell death. Recently, MOMP has been shown to be inherently pro-inflammatory with emerging cellular roles, including its ability to elicit anti-tumour immunity. Nonetheless, how MOMP triggers inflammation and how the cell regulates this remains poorly defined. We find that upon MOMP, many proteins localised either to inner or outer mitochondrial membranes are ubiquitylated in a promiscuous manner. This extensive ubiquitylation serves to recruit the essential adaptor molecule NEMO, leading to the activation of pro-inflammatory NF-κB signalling. We show that disruption of mitochondrial outer membrane integrity through different means leads to the engagement of a similar pro-inflammatory signalling platform. Therefore, mitochondrial integrity directly controls inflammation, such that permeabilised mitochondria initiate NF-κB signalling.
Keywords: Cell Death; Inflammation; Mitochondria; NF-κB; Ubiquitin.
© 2024. The Author(s).
Conflict of interest statement
SWGT consults for Exo Therapeutics. DTH consults for Triana Biomedicines. Salary support for GS was provided by a collaborative agreement with the Merck Group and Cancer Research Horizons UK. DJM has additionally received support from Puma Biotechnology and ORIC Pharmaceuticals. The remaining authors declare no competing interests.
Figures



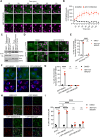


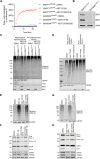
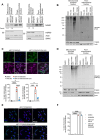
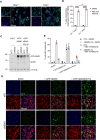

References
-
- Arganda-Carreras I, Kaynig V, Rueden C, Eliceiri KW, Schindelin J, Cardona A, Sebastian Seung H. Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification. Bioinformatics. 2017;33:2424–2426. - PubMed
-
- Bock FJ, Tait SWG. Mitochondria as multifaceted regulators of cell death. Nat Rev Mol Cell Biol. 2020;21:85–100. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

