Transcriptional programming of translation by BCL6 controls skeletal muscle proteostasis
- PMID: 38337096
- PMCID: PMC10949880
- DOI: 10.1038/s42255-024-00983-3
Transcriptional programming of translation by BCL6 controls skeletal muscle proteostasis
Abstract
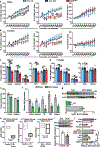
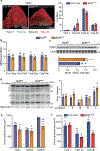
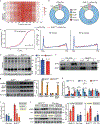
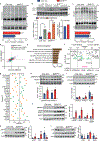
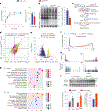
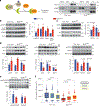
Skeletal muscle is dynamically controlled by the balance of protein synthesis and degradation. Here we discover an unexpected function for the transcriptional repressor B cell lymphoma 6 (BCL6) in muscle proteostasis and strength in mice. Skeletal muscle-specific Bcl6 ablation in utero or in adult mice results in over 30% decreased muscle mass and force production due to reduced protein synthesis and increased autophagy, while it promotes a shift to a slower myosin heavy chain fibre profile. Ribosome profiling reveals reduced overall translation efficiency in Bcl6-ablated muscles. Mechanistically, tandem chromatin immunoprecipitation, transcriptomic and translational analyses identify direct BCL6 repression of eukaryotic translation initiation factor 4E-binding protein 1 (Eif4ebp1) and activation of insulin-like growth factor 1 (Igf1) and androgen receptor (Ar). Together, these results uncover a bifunctional role for BCL6 in the transcriptional and translational control of muscle proteostasis.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures

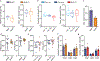






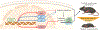
References
-
- Cohen S, Nathan JA & Goldberg AL Muscle wasting in disease: molecular mechanisms and promising therapies. Nat. Rev. Drug Discov. 14, 58–74 (2015). - PubMed
-
- Saxton RA & Sabatini DM mTOR signaling in growth, metabolism, and disease. Cell 169, 361–371 (2017). - PubMed
-
- Bodine SC et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science 294, 1704–1708 (2001). - PubMed
-
- Brunet A et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857–868 (1999). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

