CircRNAome of Childhood Acute Lymphoblastic Leukemia: Deciphering Subtype-Specific Expression Profiles and Involvement in TCF3::PBX1 ALL
- PMID: 38338754
- PMCID: PMC10855129
- DOI: 10.3390/ijms25031477
CircRNAome of Childhood Acute Lymphoblastic Leukemia: Deciphering Subtype-Specific Expression Profiles and Involvement in TCF3::PBX1 ALL
Abstract
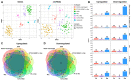
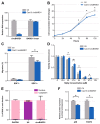
Childhood B-cell acute lymphoblastic leukemia (B-ALL) is a heterogeneous disease comprising multiple molecular subgroups with subtype-specific expression profiles. Recently, a new type of ncRNA, termed circular RNA (circRNA), has emerged as a promising biomarker in cancer, but little is known about their role in childhood B-ALL. Here, through RNA-seq analysis in 105 childhood B-ALL patients comprising six genetic subtypes and seven B-cell controls from two independent cohorts we demonstrated that circRNAs properly stratified B-ALL subtypes. By differential expression analysis of each subtype vs. controls, 156 overexpressed and 134 underexpressed circRNAs were identified consistently in at least one subtype, most of them with subtype-specific expression. TCF3::PBX1 subtype was the one with the highest number of unique and overexpressed circRNAs, and the circRNA signature could effectively discriminate new patients with TCF3::PBX1 subtype from others. Our results indicated that NUDT21, an RNA-binding protein (RBP) involved in circRNA biogenesis, may contribute to this circRNA enrichment in TCF3::PBX1 ALL. Further functional characterization using the CRISPR-Cas13d system demonstrated that circBARD1, overexpressed in TCF3::PBX1 patients and regulated by NUDT21, might be involved in leukemogenesis through the activation of p38 via hsa-miR-153-5p. Our results suggest that circRNAs could play a role in the pathogenesis of childhood B-ALL.
Keywords: NUDT21; RNA-binding protein; TCF3::PBX1; acute lymphoblastic leukemia; childhood B-ALL; circular RNA.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- Lajoie M., Drouin S., Caron M., St-Onge P., Ouimet M., Gioia R., Lafond M.H., Vidal R., Richer C., Oualkacha K., et al. Specific expression of novel long non-coding RNAs in high-hyperdiploid childhood acute lymphoblastic leukemia. PLoS ONE. 2017;12:e0174124. doi: 10.1371/journal.pone.0174124. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

