Differential Gene Expression following DHX36/ G4R1 Knockout Is Associated with G-Quadruplex Content and Cancer
- PMID: 38339029
- PMCID: PMC10855491
- DOI: 10.3390/ijms25031753
Differential Gene Expression following DHX36/ G4R1 Knockout Is Associated with G-Quadruplex Content and Cancer
Abstract
G-quadruplexes (G4s) are secondary DNA and RNA structures stabilized by positive cations in a central channel formed by stacked tetrads of Hoogsteen base-paired guanines. G4s form from G-rich sequences across the genome, whose biased distribution in regulatory regions points towards a gene-regulatory role. G4s can themselves be regulated by helicases, such as DHX36 (aliases: G4R1 and RHAU), which possess the necessary activity to resolve these stable structures. G4s have been shown to both positively and negatively regulate gene expression when stabilized by ligands, or through the loss of helicase activity. Using DHX36 knockout Jurkat cell lines, we identified widespread, although often subtle, effects on gene expression that are associated with the presence or number of observed G-quadruplexes in promoters or gene regions. Genes that significantly change their expression, particularly those that show a significant increase in RNA abundance under DHX36 knockout, are associated with a range of cellular functions and processes, including numerous transcription factors and oncogenes, and are linked to several cancers. Our work highlights the direct and indirect role of DHX36 in the transcriptome of T-lymphocyte leukemia cells and the potential for DHX36 dysregulation in cancer.
Keywords: DHX36; G-quadruplex; G4R1; Jurkat cells; RHAU; helicase; leukemia.
Conflict of interest statement
Author James P. Vaughn was employed by the company NanoMedica LLC. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
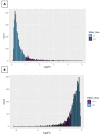
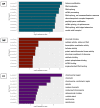

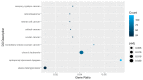
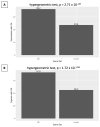

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

