AAV-RPGR Gene Therapy Rescues Opsin Mislocalisation in a Human Retinal Organoid Model of RPGR-Associated X-Linked Retinitis Pigmentosa
- PMID: 38339118
- PMCID: PMC10855600
- DOI: 10.3390/ijms25031839
AAV-RPGR Gene Therapy Rescues Opsin Mislocalisation in a Human Retinal Organoid Model of RPGR-Associated X-Linked Retinitis Pigmentosa
Abstract
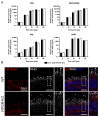
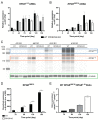
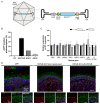
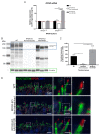
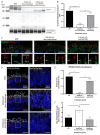
Variants within the Retinitis Pigmentosa GTPase regulator (RPGR) gene are the predominant cause of X-Linked Retinitis Pigmentosa (XLRP), a common and severe form of inherited retinal disease. XLRP is characterised by the progressive degeneration and loss of photoreceptors, leading to visual loss and, ultimately, bilateral blindness. Unfortunately, there are no effective approved treatments for RPGR-associated XLRP. We sought to investigate the efficacy of RPGRORF15 gene supplementation using a clinically relevant construct in human RPGR-deficient retinal organoids (ROs). Isogenic RPGR knockout (KO)-induced pluripotent stem cells (IPSCs) were generated using established CRISPR/Cas9 gene editing methods targeting RPGR. RPGR-KO and isogenic wild-type IPSCs were differentiated into ROs and utilised to test the adeno associated virus (AAV) RPGR (AAV-RPGR) clinical vector construct. The transduction of RPGR-KO ROs using AAV-RPGR successfully restored RPGR mRNA and protein expression and localisation to the photoreceptor connecting cilium in rod and cone photoreceptors. Vector-derived RPGR demonstrated equivalent levels of glutamylation to WT ROs. In addition, treatment with AAV-RPGR restored rhodopsin localisation within RPGR-KO ROs, reducing mislocalisation to the photoreceptor outer nuclear layer. These data provide mechanistic insights into RPGRORF15 gene supplementation functional potency in human photoreceptor cells and support the previously reported Phase I/II trial positive results using this vector construct in patients with RPGR-associated XLRP, which is currently being tested in a Phase III clinical trial.
Keywords: CRISPR/Cas9; IPSC; RPGR; X-linked; adeno associated virus; gene therapy; retinal organoids; retinitis pigmentosa.
Conflict of interest statement
All authors are affiliated with MeiraGTx. P.E.S., A.N., T.A-I., T.V., S.L.B., S.N., A.F., A.L. and A.G. are employees of MeiraGTx. M.M. provides consultancy services to MeiraGTx. The authors have no other relevant affiliations or financial involvement with any organisation or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Figures
References
-
- Meindl A., Dry K., Herrmann K., Manson F., Ciccodicola A., Edgar A., Carvalho M.R.S., Achatz H., Hellebrand H., Lennon A., et al. A Gene (RPGR) with Homology to the RCC1 Guanine Nucleotide Exchange Factor Is Mutated in X-Linked Retinitis Pigmentosa (RP3) Nat. Genet. 1996;13:35–42. doi: 10.1038/ng0596-35. - DOI - PubMed
-
- Jin Z.B., Liu X.Q., Hayakawa M., Murakami A., Nao-i N. Mutational Analysis of RPGR and RP2 Genes in Japanese Patients with Retinitis Pigmentosa: Identification of Four Mutations. Mol. Vis. 2006;12:1167–1174. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

