Transcription factor PagMYB31 positively regulates cambium activity and negatively regulates xylem development in poplar
- PMID: 38339982
- PMCID: PMC11062435
- DOI: 10.1093/plcell/koae040
Transcription factor PagMYB31 positively regulates cambium activity and negatively regulates xylem development in poplar
Abstract
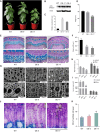
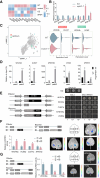
Wood formation involves consecutive developmental steps, including cell division of vascular cambium, xylem cell expansion, secondary cell wall (SCW) deposition, and programmed cell death. In this study, we identified PagMYB31 as a coordinator regulating these processes in Populus alba × Populus glandulosa and built a PagMYB31-mediated transcriptional regulatory network. PagMYB31 mutation caused fewer layers of cambial cells, larger fusiform initials, ray initials, vessels, fiber and ray cells, and enhanced xylem cell SCW thickening, showing that PagMYB31 positively regulates cambial cell proliferation and negatively regulates xylem cell expansion and SCW biosynthesis. PagMYB31 repressed xylem cell expansion and SCW thickening through directly inhibiting wall-modifying enzyme genes and the transcription factor genes that activate the whole SCW biosynthetic program, respectively. In cambium, PagMYB31 could promote cambial activity through TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR (TDIF)/PHLOEM INTERCALATED WITH XYLEM (PXY) signaling by directly regulating CLAVATA3/ESR-RELATED (CLE) genes, and it could also directly activate WUSCHEL HOMEOBOX RELATED4 (PagWOX4), forming a feedforward regulation. We also observed that PagMYB31 could either promote cell proliferation through the MYB31-MYB72-WOX4 module or inhibit cambial activity through the MYB31-MYB72-VASCULAR CAMBIUM-RELATED MADS2 (VCM2)/PIN-FORMED5 (PIN5) modules, suggesting its role in maintaining the homeostasis of vascular cambium. PagMYB31 could be a potential target to manipulate different developmental stages of wood formation.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. The authors declare no competing interests.
Figures





References
-
- Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, Borrega N, Hervé J, Blondet E, Balzergue S, et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell. 2011:23(3):1124–1137. 10.1105/tpc.110.082792 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

