Tumoral and stromal hMENA isoforms impact tertiary lymphoid structure localization in lung cancer and predict immune checkpoint blockade response in patients with cancer
- PMID: 38340557
- PMCID: PMC10869748
- DOI: 10.1016/j.ebiom.2024.105003
Tumoral and stromal hMENA isoforms impact tertiary lymphoid structure localization in lung cancer and predict immune checkpoint blockade response in patients with cancer
Abstract
Background: Tertiary Lymphoid Structures (TLS) correlate with positive outcomes in patients with NSCLC and the efficacy of immune checkpoint blockade (ICB) in cancer. The actin regulatory protein hMENA undergoes tissue-specific splicing, producing the epithelial hMENA11a linked to favorable prognosis in early NSCLC, and the mesenchymal hMENAΔv6 found in invasive cancer cells and pro-tumoral cancer-associated fibroblasts (CAFs). This study investigates how hMENA isoforms in tumor cells and CAFs relate to TLS presence, localization and impact on patient outcomes and ICB response.
Methods: Methods involved RNA-SEQ on NSCLC cells with depleted hMENA isoforms. A retrospective observational study assessed tissues from surgically treated N0 patients with NSCLC, using immunohistochemistry for tumoral and stromal hMENA isoforms, fibronectin, and TLS presence. ICB-treated patient tumors were analyzed using Nanostring nCounter and GeoMx spatial transcriptomics. Multiparametric flow cytometry characterized B cells and tissue-resident memory T cells (TRM). Survival and ICB response were estimated in the cohort and validated using bioinformatics pipelines in different datasets.
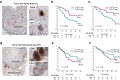
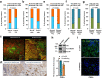
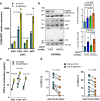
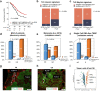
Findings: Findings indicate that hMENA11a in NSCLC cells upregulates the TLS regulator LTβR, decreases fibronectin, and favors CXCL13 production by TRM. Conversely, hMENAΔv6 in CAFs inhibits LTβR-related NF-kB pathway, reduces CXCL13 secretion, and promotes fibronectin production. These patterns are validated in N0 NSCLC tumors, where hMENA11ahigh expression, CAF hMENAΔv6low, and stromal fibronectinlow are associated with intratumoral TLS, linked to memory B cells and predictive of longer survival. The hMENA isoform pattern, fibronectin, and LTβR expression broadly predict ICB response in tumors where TLS indicates an anti-tumor immune response.
Interpretation: This study uncovers hMENA alternative splicing as an unexplored contributor to TLS-related Tumor Immune Microenvironment (TIME) and a promising biomarker for clinical outcomes and likely ICB responsiveness in N0 patients with NSCLC.
Funding: This work is supported by AIRC (IG 19822), ACC (RCR-2019-23669120), CAL.HUB.RIA Ministero Salute PNRR-POS T4, "Ricerca Corrente" granted by the Italian Ministry of Health.
Keywords: Cancer-associated fibroblasts; Epithelial mesenchymal transition; Fibronectin; Immune checkpoint blockade; Non-small cell lung cancer; Resistance to immunotherapy; Tertiary lymphoid structures; Tumor microenvironment; hMENA isoforms.
Copyright © 2024 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests F.D.M. and P.N. are inventors of patents on the role of hMENA isoforms on tumor progression and response to therapies. S.W. was an employee and stockholder of NanoString Technologies at the time the work was performed. The other authors declare no competing interests.
Figures


References
-
- Reck M., Rodríguez-Abreu D., Robinson A.G., et al. Updated analysis of KEYNOTE-024: pembrolizumab versus platinum-based chemotherapy for advanced non-small-cell lung cancer with PD-L1 tumor proportion score of 50% or greater. J Clin Oncol. 2019;37(7):537–546. - PubMed
-
- Martinez P., Peters S., Stammers T., Soria J.C. Immunotherapy for the first-line treatment of patients with metastatic non-small cell lung cancer. Clin Cancer Res. 2019;25(9):2691–2698. - PubMed
-
- Galon J., Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov. 2019;18(3):197–218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

