An enhanced broad-spectrum peptide inhibits Omicron variants in vivo
- PMID: 38340726
- PMCID: PMC10897629
- DOI: 10.1016/j.xcrm.2024.101418
An enhanced broad-spectrum peptide inhibits Omicron variants in vivo
Abstract
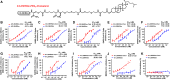
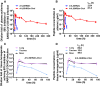
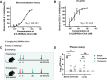
The continual emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs) poses a major challenge to vaccines and antiviral therapeutics due to their extensive evasion of immunity. Aiming to develop potent and broad-spectrum anticoronavirus inhibitors, we generated A1-(GGGGS)7-HR2m (A1L35HR2m) by introducing an angiotensin-converting enzyme 2 (ACE2)-derived peptide A1 to the N terminus of the viral HR2-derived peptide HR2m through a long flexible linker, which showed significantly improved antiviral activity. Further cholesterol (Chol) modification at the C terminus of A1L35HR2m greatly enhanced the inhibitory activities against SARS-CoV-2, SARS-CoV-2 VOCs, SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV) pseudoviruses, with IC50 values ranging from 0.16 to 5.53 nM. A1L35HR2m-Chol also potently inhibits spike-protein-mediated cell-cell fusion and the replication of authentic Omicron BA.2.12.1, BA.5, and EG.5.1. Importantly, A1L35HR2m-Chol distributed widely in respiratory tract tissue and had a long half-life (>10 h) in vivo. Intranasal administration of A1L35HR2m-Chol to K18-hACE2 transgenic mice potently inhibited Omicron BA.5 and EG.5.1 infection both prophylactically and therapeutically.
Keywords: ACE2 peptide; HR2 peptide; SARS-CoV-2; cholesterol modification; lipopeptide; pan-coronavirus fusion inhibitor; synergistic effect; variants of concern.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests B.D., W.B., S.Y., K.T., and G.C. are the inventors of a provisional patent filed by Westlake University and The University of Hong Kong.
Figures





References
-
- Kuzmina A., Khalaila Y., Voloshin O., Keren-Naus A., Boehm-Cohen L., Raviv Y., Shemer-Avni Y., Rosenberg E., Taube R. SARS-CoV-2 spike variants exhibit differential infectivity and neutralization resistance to convalescent or post-vaccination sera. Cell Host Microbe. 2021;29:522–528.e2. doi: 10.1016/j.chom.2021.03.008. - DOI - PMC - PubMed
-
- Garcia-Beltran W.F., Lam E.C., St Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2523. doi: 10.1016/j.cell.2021.04.006. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

