Lysyl Oxidase Regulates Epithelial Differentiation and Barrier Integrity in Eosinophilic Esophagitis
- PMID: 38340809
- PMCID: PMC11026689
- DOI: 10.1016/j.jcmgh.2024.01.025
Lysyl Oxidase Regulates Epithelial Differentiation and Barrier Integrity in Eosinophilic Esophagitis
Abstract
Background & aims: Epithelial disruption in eosinophilic esophagitis (EoE) encompasses both impaired differentiation and diminished barrier integrity. We have shown that lysyl oxidase (LOX), a collagen cross-linking enzyme, is up-regulated in the esophageal epithelium in EoE. However, the functional roles of LOX in the esophageal epithelium remains unknown.
Methods: We investigated roles for LOX in the human esophageal epithelium using 3-dimensional organoid and air-liquid interface cultures stimulated with interleukin (IL)13 to recapitulate the EoE inflammatory milieu, followed by single-cell RNA sequencing, quantitative reverse-transcription polymerase chain reaction, Western blot, histology, and functional analyses of barrier integrity.
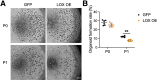
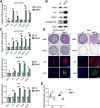
Results: Single-cell RNA sequencing analysis on patient-derived organoids revealed that LOX was induced by IL13 in differentiated cells. LOX-overexpressing organoids showed suppressed basal and up-regulated differentiation markers. In addition, LOX overexpression enhanced junctional protein genes and transepithelial electrical resistance. LOX overexpression restored the impaired differentiation and barrier function, including in the setting of IL13 stimulation. Transcriptome analyses on LOX-overexpressing organoids identified an enriched bone morphogenetic protein (BMP) signaling pathway compared with wild-type organoids. In particular, LOX overexpression increased BMP2 and decreased the BMP antagonist follistatin. Finally, we found that BMP2 treatment restored the balance of basal and differentiated cells.
Conclusions: Our data support a model whereby LOX exhibits noncanonical roles as a signaling molecule important for epithelial homeostasis in the setting of inflammation via activation of the BMP pathway in the esophagus. The LOX/BMP axis may be integral in esophageal epithelial differentiation and a promising target for future therapies.
Keywords: BMP; Eosinophilic Esophagitis; Lysyl Oxidase; Organoid.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





Update of
-
Lysyl oxidase regulates epithelial differentiation and barrier integrity in eosinophilic esophagitis.bioRxiv [Preprint]. 2023 Mar 28:2023.03.27.534387. doi: 10.1101/2023.03.27.534387. bioRxiv. 2023. Update in: Cell Mol Gastroenterol Hepatol. 2024;17(6):923-937. doi: 10.1016/j.jcmgh.2024.01.025. PMID: 37034590 Free PMC article. Updated. Preprint.
References
-
- Chen W., Yang A., Jia J., et al. Lysyl oxidase (LOX) family members: rationale and their potential as therapeutic targets for liver fibrosis. Nat Rev Gastroenterol Hepatol. 2020;72:729–741. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

