Fully automated whole brain segmentation from rat MRI scans with a convolutional neural network
- PMID: 38340902
- PMCID: PMC11000587
- DOI: 10.1016/j.jneumeth.2024.110078
Fully automated whole brain segmentation from rat MRI scans with a convolutional neural network
Abstract
Background: Whole brain delineation (WBD) is utilized in neuroimaging analysis for data preprocessing and deriving whole brain image metrics. Current automated WBD techniques for analysis of preclinical brain MRI data show limited accuracy when images present with significant neuropathology and anatomical deformations, such as that resulting from organophosphate intoxication (OPI) and Alzheimer's Disease (AD), and inadequate generalizability.
Methods: A modified 2D U-Net framework was employed for WBD of MRI rodent brains, consisting of 27 convolutional layers, batch normalization, two dropout layers and data augmentation, after training parameter optimization. A total of 265 T2-weighted 7.0 T MRI scans were utilized for the study, including 125 scans of an OPI rat model for neural network training. For testing and validation, 20 OPI rat scans and 120 scans of an AD rat model were utilized. U-Net performance was evaluated using Dice coefficients (DC) and Hausdorff distances (HD) between the U-Net-generated and manually segmented WBDs.
Results: The U-Net achieved a DC (median[range]) of 0.984[0.936-0.990] and HD of 1.69[1.01-6.78] mm for OPI rat model scans, and a DC (mean[range]) of 0.975[0.898-0.991] and HD of 1.49[0.86-3.89] for the AD rat model scans.
Comparison with existing methods: The proposed approach is fully automated and robust across two rat strains and longitudinal brain changes with a computational speed of 8 seconds/scan, overcoming limitations of manual segmentation.
Conclusions: The modified 2D U-Net provided a fully automated, efficient, and generalizable segmentation approach that achieved high accuracy across two disparate rat models of neurological diseases.
Keywords: Automated Segmentation; MRI; Machine Learning; Preclinical Neuroimaging; Rodent Brain Imaging; Skull Stripping.
Copyright © 2024 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest None of the authors have a conflict of interest with the work presented.
Figures

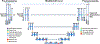
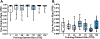
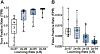





Similar articles
-
Three-dimensional U-Net with transfer learning improves automated whole brain delineation from MRI brain scans of rats, mice, and monkeys.Comput Biol Med. 2025 Sep;195:110569. doi: 10.1016/j.compbiomed.2025.110569. Epub 2025 Jun 20. Comput Biol Med. 2025. PMID: 40543276
-
Study of multistep Dense U-Net-based automatic segmentation for head MRI scans.Med Phys. 2024 Mar;51(3):2230-2238. doi: 10.1002/mp.16824. Epub 2023 Nov 13. Med Phys. 2024. PMID: 37956307
-
Automated joint skull-stripping and segmentation with Multi-Task U-Net in large mouse brain MRI databases.Neuroimage. 2021 Apr 1;229:117734. doi: 10.1016/j.neuroimage.2021.117734. Epub 2021 Jan 14. Neuroimage. 2021. PMID: 33454412
-
A generalizable brain extraction net (BEN) for multimodal MRI data from rodents, nonhuman primates, and humans.Elife. 2022 Dec 22;11:e81217. doi: 10.7554/eLife.81217. Elife. 2022. PMID: 36546674 Free PMC article.
-
RU-Net: skull stripping in rat brain MR images after ischemic stroke with rat U-Net.BMC Med Imaging. 2023 Mar 27;23(1):44. doi: 10.1186/s12880-023-00994-8. BMC Med Imaging. 2023. PMID: 36973775 Free PMC article.
Cited by
-
Three-dimensional U-Net with transfer learning improves automated whole brain delineation from MRI brain scans of rats, mice, and monkeys.Comput Biol Med. 2025 Sep;195:110569. doi: 10.1016/j.compbiomed.2025.110569. Epub 2025 Jun 20. Comput Biol Med. 2025. PMID: 40543276
-
Shifts in the spatiotemporal profile of inflammatory phenotypes of innate immune cells in the rat brain following acute intoxication with the organophosphate diisopropylfluorophosphate.J Neuroinflammation. 2024 Nov 4;21(1):285. doi: 10.1186/s12974-024-03272-8. J Neuroinflammation. 2024. PMID: 39497181 Free PMC article.
References
-
- Almeida AJD, Hobson BA, Saito N, Harvey D, Bruun DA, Porter VA, Garbow JR, Chaudhari AJ, Lein PJ, Quantitative T2 mapping-based longitudinal assessment of brain injury and therapeutic rescue in the rat following acute organophosphate intoxication. Manuscript submitted for publication. - PMC - PubMed
-
- Azad R, Aghdam EK, Rauland A, Jia Y, Avval AH, Bozorgpour A, Karimijafarbigloo S, Cohen JP, Adeli E, Merhof D, 2022. Medical Image Segmentation Review: The success of U-Net. - PubMed

