Tregs from human blood differentiate into nonlymphoid tissue-resident effector cells upon TNFR2 costimulation
- PMID: 38341270
- PMCID: PMC10972588
- DOI: 10.1172/jci.insight.172942
Tregs from human blood differentiate into nonlymphoid tissue-resident effector cells upon TNFR2 costimulation
Abstract
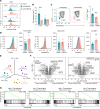
Tregs can facilitate transplant tolerance and attenuate autoimmune and inflammatory diseases. Therefore, it is clinically relevant to stimulate Treg expansion and function in vivo and to create therapeutic Treg products in vitro. We report that TNF receptor 2 (TNFR2) is a unique costimulus for naive, thymus-derived Tregs (tTregs) from human blood that promotes their differentiation into nonlymphoid tissue-resident (NLT-resident) effector Tregs, without Th-like polarization. In contrast, CD28 costimulation maintains a lymphoid tissue-resident (LT-resident) Treg phenotype. We base this conclusion on transcriptome and proteome analysis of TNFR2- and CD28-costimulated CD4+ tTregs and conventional T cells (Tconvs), followed by bioinformatic comparison with published transcriptomic Treg signatures from NLT and LT in health and disease, including autoimmunity and cancer. These analyses illuminate that TNFR2 costimulation promoted tTreg capacity for survival, migration, immunosuppression, and tissue regeneration. Functional studies confirmed improved migratory ability of TNFR2-costimulated tTregs. Flow cytometry validated the presence of the TNFR2-driven tTreg signature in effector/memory Tregs from the human placenta, as opposed to blood. Thus, TNFR2 can be exploited as a driver of NLT-resident tTreg differentiation for adoptive cell therapy or antibody-based immunomodulation in human disease.
Keywords: Bioinformatics; Costimulation; Immunology; T cells.
Conflict of interest statement
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

