Integrated multi-omic analysis identifies fatty acid binding protein 4 as a biomarker and therapeutic target of ischemia-reperfusion injury in steatotic liver transplantation
- PMID: 38341383
- PMCID: PMC10858962
- DOI: 10.1007/s00018-023-05110-1
Integrated multi-omic analysis identifies fatty acid binding protein 4 as a biomarker and therapeutic target of ischemia-reperfusion injury in steatotic liver transplantation
Abstract
Background and aims: Due to a lack of donor grafts, steatotic livers are used more often for liver transplantation (LT). However, steatotic donor livers are more sensitive to ischemia-reperfusion (IR) injury and have a worse prognosis after LT. Efforts to optimize steatotic liver grafts by identifying injury targets and interventions have become a hot issue.
Methods: Mouse LT models were established, and 4D label-free proteome sequencing was performed for four groups: normal control (NC) SHAM, high-fat (HF) SHAM, NC LT, and HF LT to screen molecular targets for aggravating liver injury in steatotic LT. Expression detection of molecular targets was performed based on liver specimens from 110 donors to verify its impact on the overall survival of recipients. Pharmacological intervention using small-molecule inhibitors on an injury-related target was used to evaluate the therapeutic effect. Transcriptomics and metabolomics were performed to explore the regulatory network and further integrated bioinformatics analysis and multiplex immunofluorescence were adopted to assess the regulation of pathways and organelles.
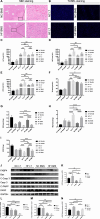
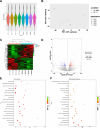
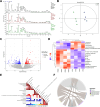
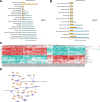
Results: HF LT group represented worse liver function compared with NC LT group, including more apoptotic hepatocytes (P < 0.01) and higher serum transaminase (P < 0.05). Proteomic results revealed that the mitochondrial membrane, endocytosis, and oxidative phosphorylation pathways were upregulated in HF LT group. Fatty acid binding protein 4 (FABP4) was identified as a hypoxia-inducible protein (fold change > 2 and P < 0.05) that sensitized mice to IR injury in steatotic LT. The overall survival of recipients using liver grafts with high expression of FABP4 was significantly worse than low expression of FABP4 (68.5 vs. 87.3%, P < 0.05). Adoption of FABP4 inhibitor could protect the steatotic liver from IR injury during transplantation, including reducing hepatocyte apoptosis, reducing serum transaminase (P < 0.05), and alleviating oxidative stress damage (P < 0.01). According to integrated transcriptomics and metabolomics analysis, cAMP signaling pathway was enriched following FABP4 inhibitor use. The activation of cAMP signaling pathway was validated. Microscopy and immunofluorescence staining results suggested that FABP4 inhibitors could regulate mitochondrial membrane homeostasis in steatotic LT.
Conclusions: FABP4 was identified as a hypoxia-inducible protein that sensitized steatotic liver grafts to IR injury. The FABP4 inhibitor, BMS-309403, could activate of cAMP signaling pathway thereby modulating mitochondrial membrane homeostasis, reducing oxidative stress injury in steatotic donors.
Keywords: Fatty acid binding protein 4; Liver steatosis; Liver transplantation; Metabolomics; Mitochondrion; Proteomics; Transcriptomics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest related to this study.
Figures






Similar articles
-
Fatty acid binding protein-4 (FABP4) is a hypoxia inducible gene that sensitizes mice to liver ischemia/reperfusion injury.J Hepatol. 2015 Oct;63(4):855-62. doi: 10.1016/j.jhep.2015.05.030. Epub 2015 Jun 10. J Hepatol. 2015. PMID: 26070408 Free PMC article.
-
Attenuation of Ischemia-Reperfusion Injury and Improvement of Survival in Recipients of Steatotic Rat Livers Using CD47 Monoclonal Antibody.Transplantation. 2016 Jul;100(7):1480-9. doi: 10.1097/TP.0000000000001186. Transplantation. 2016. PMID: 27331362
-
NO-IL-6/10-IL-1β axis: a new pathway in steatotic and non-steatotic liver grafts from brain-dead donor rats.Front Immunol. 2023 Aug 1;14:1178909. doi: 10.3389/fimmu.2023.1178909. eCollection 2023. Front Immunol. 2023. PMID: 37593740 Free PMC article.
-
Adipocytokines in Steatotic Liver Surgery/Transplantation.Transplantation. 2019 Jan;103(1):71-77. doi: 10.1097/TP.0000000000002098. Transplantation. 2019. PMID: 30586349 Review.
-
Ischemia–reperfusion injury in patients with fatty liver and the clinical impact of steatotic liver on hepatic surgery.Surg Today. 2014 Sep;44(9):1611-25. doi: 10.1007/s00595-013-0736-9. Surg Today. 2014. PMID: 24078000 Review.
Cited by
-
Molecular Mechanisms of Ischemia/Reperfusion Injury and Graft Dysfunction in Liver Transplantation: Insights from Multi-Omics Studies in Rodent Animal Models.Int J Biol Sci. 2025 Feb 24;21(5):2135-2154. doi: 10.7150/ijbs.109449. eCollection 2025. Int J Biol Sci. 2025. PMID: 40083684 Free PMC article. Review.
-
Olaparib promotes FABP4 expression and reduces antitumor effect in ovarian cancer cells with a BRCA1 mutation.Oncol Lett. 2024 Nov 20;29(2):67. doi: 10.3892/ol.2024.14813. eCollection 2025 Feb. Oncol Lett. 2024. PMID: 39619421 Free PMC article.
-
Schematic Assessment of Metabolic Signatures of Non-alcoholic Fatty Liver Disease by Bridging Endocrinology and Internal Medicine: A Precision Therapy-Based Meta-Analysis.Cureus. 2025 Apr 28;17(4):e83133. doi: 10.7759/cureus.83133. eCollection 2025 Apr. Cureus. 2025. PMID: 40438852 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
- No. 2021YFA1100500/National Key Research and Development Program of China
- ZR2023QH315/Youth Program of Natural Science Foundation of Shandong
- 81625003/National Natural Science Funds for Distinguished Young Scholar of China
- 81930016/National Natural Science Foundation of China
- LQ17H160006/Youth Program of Natural Science Foundation of Zhejiang Province
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

