1,4-dihydroxy quininib activates ferroptosis pathways in metastatic uveal melanoma and reveals a novel prognostic biomarker signature
- PMID: 38341410
- PMCID: PMC10858877
- DOI: 10.1038/s41420-023-01773-8
1,4-dihydroxy quininib activates ferroptosis pathways in metastatic uveal melanoma and reveals a novel prognostic biomarker signature
Abstract
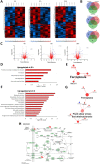
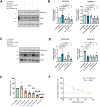
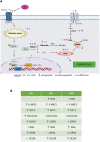
Uveal melanoma (UM) is an ocular cancer, with propensity for lethal liver metastases. When metastatic UM (MUM) occurs, as few as 8% of patients survive beyond two years. Efficacious treatments for MUM are urgently needed. 1,4-dihydroxy quininib, a cysteinyl leukotriene receptor 1 (CysLT1) antagonist, alters UM cancer hallmarks in vitro, ex vivo and in vivo. Here, we investigated the 1,4-dihydroxy quininib mechanism of action and its translational potential in MUM. Proteomic profiling of OMM2.5 cells identified proteins differentially expressed after 1,4-dihydroxy quininib treatment. Glutathione peroxidase 4 (GPX4), glutamate-cysteine ligase modifier subunit (GCLM), heme oxygenase 1 (HO-1) and 4 hydroxynonenal (4-HNE) expression were assessed by immunoblots. Biliverdin, glutathione and lipid hydroperoxide were measured biochemically. Association between the expression of a specific ferroptosis signature and UM patient survival was performed using public databases. Our data revealed that 1,4-dihydroxy quininib modulates the expression of ferroptosis markers in OMM2.5 cells. Biochemical assays validated that GPX4, biliverdin, GCLM, glutathione and lipid hydroperoxide were significantly altered. HO-1 and 4-HNE levels were significantly increased in MUM tumor explants from orthotopic patient-derived xenografts (OPDX). Expression of genes inhibiting ferroptosis is significantly increased in UM patients with chromosome 3 monosomy. We identified IFerr, a novel ferroptosis signature correlating with UM patient survival. Altogether, we demontrated that in MUM cells and tissues, 1,4-dihydroxy quininib modulates key markers that induce ferroptosis, a relatively new type of cell death driven by iron-dependent peroxidation of phospholipids. Furthermore, we showed that high expression of specific genes inhibiting ferroptosis is associated with a worse UM prognosis, thus, the IFerr signature is a potential prognosticator for which patients develop MUM. All in all, ferroptosis has potential as a clinical biomarker and therapeutic target for MUM.
© 2024. The Author(s).
Conflict of interest statement
VT, EOR, KS and BNK are named inventors on a filed patent application related to UM biomarkers. AV is co-founder of Xenopat S.L.
Figures





Similar articles
-
Assessment of ferroptosis as a promising candidate for metastatic uveal melanoma treatment and prognostication.Front Pharmacol. 2024 Oct 1;15:1466896. doi: 10.3389/fphar.2024.1466896. eCollection 2024. Front Pharmacol. 2024. PMID: 39411069 Free PMC article. Review.
-
1,4-dihydroxy quininib modulates the secretome of uveal melanoma tumour explants and a marker of oxidative phosphorylation in a metastatic xenograft model.Front Med (Lausanne). 2023 Jan 9;9:1036322. doi: 10.3389/fmed.2022.1036322. eCollection 2022. Front Med (Lausanne). 2023. PMID: 36698840 Free PMC article.
-
High Cysteinyl Leukotriene Receptor 1 Expression Correlates with Poor Survival of Uveal Melanoma Patients and Cognate Antagonist Drugs Modulate the Growth, Cancer Secretome, and Metabolism of Uveal Melanoma Cells.Cancers (Basel). 2020 Oct 13;12(10):2950. doi: 10.3390/cancers12102950. Cancers (Basel). 2020. PMID: 33066024 Free PMC article.
-
Ergolide mediates anti-cancer effects on metastatic uveal melanoma cells and modulates their cellular and extracellular vesicle proteomes.Open Res Eur. 2023 Nov 13;3:88. doi: 10.12688/openreseurope.15973.2. eCollection 2023. Open Res Eur. 2023. PMID: 37981907 Free PMC article.
-
Lipid Peroxidation-Dependent Cell Death Regulated by GPx4 and Ferroptosis.Curr Top Microbiol Immunol. 2017;403:143-170. doi: 10.1007/82_2016_508. Curr Top Microbiol Immunol. 2017. PMID: 28204974 Review.
Cited by
-
The role of signaling pathways mediated by the GPCRs CysLTR1/2 in melanocyte proliferation and senescence.Sci Signal. 2024 Sep 17;17(854):eadp3967. doi: 10.1126/scisignal.adp3967. Epub 2024 Sep 17. Sci Signal. 2024. PMID: 39288219 Review.
-
Global trends and emerging insights in ocular tumor-associated metabolites: a bibliometric and visualization analysis.Front Mol Biosci. 2025 Jun 18;12:1572710. doi: 10.3389/fmolb.2025.1572710. eCollection 2025. Front Mol Biosci. 2025. PMID: 40607060 Free PMC article.
-
Haloperidol drug repurposing unveils ferroptosis involvement in breast cancer cells.Sci Rep. 2025 Jul 24;15(1):26948. doi: 10.1038/s41598-025-12645-9. Sci Rep. 2025. PMID: 40707640 Free PMC article.
-
Assessment of ferroptosis as a promising candidate for metastatic uveal melanoma treatment and prognostication.Front Pharmacol. 2024 Oct 1;15:1466896. doi: 10.3389/fphar.2024.1466896. eCollection 2024. Front Pharmacol. 2024. PMID: 39411069 Free PMC article. Review.
-
The HELP-UnaG Fusion Protein as a Bilirubin Biosensor: From Theory to Mature Technological Development.Molecules. 2025 Jan 21;30(3):439. doi: 10.3390/molecules30030439. Molecules. 2025. PMID: 39942546 Free PMC article. Review.
References
-
- Dogrusöz M, Jager MJ, Damato B. Uveal Melanoma Treatment and Prognostication. Asia-Pacific J Ophthalmol. 2017;6:186. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

