Hsa_circ_0021205 enhances lipolysis via regulating miR-195-5p/HSL axis and drives malignant progression of glioblastoma
- PMID: 38341418
- PMCID: PMC10858904
- DOI: 10.1038/s41420-024-01841-7
Hsa_circ_0021205 enhances lipolysis via regulating miR-195-5p/HSL axis and drives malignant progression of glioblastoma
Abstract
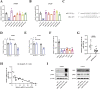
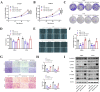
Abnormal lipid metabolism is an essential hallmark of glioblastoma. Hormone sensitive lipase (HSL), an important rate-limiting enzyme contributed to lipolysis, which was involved in aberrant lipolysis of glioblastoma, however, its definite roles and the relevant regulatory pathway have not been fully elucidated. Our investigations disclosed high expression of HSL in glioblastoma. Knock-down of HSL restrained proliferation, migration, and invasion of glioblastoma cells while adding to FAs could significantly rescue the inhibitory effect of si-HSL on tumor cells. Overexpression of HSL further promoted tumor cell proliferation and invasion. Bioinformatics analysis and dual-luciferase reporter assay were performed to predict and verify the regulatory role of ncRNAs on HSL. Mechanistically, hsa_circ_0021205 regulated HSL expression by sponging miR-195-5p, which further promoted lipolysis and drove the malignant progression of glioblastoma. Besides, hsa_circ_0021205/miR-195-5p/HSL axis activated the epithelial-mesenchymal transition (EMT) signaling pathway. These findings suggested that hsa_circ_0021205 promoted tumorigenesis of glioblastoma through regulation of HSL, and targeting hsa_circ_0021205/miR-195-5p/HSL axis can serve as a promising new strategy against glioblastoma.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Blocking hsa_circ_0006168 suppresses cell proliferation and motility of human glioblastoma cells by regulating hsa_circ_0006168/miR-628-5p/IGF1R ceRNA axis.Cell Cycle. 2021 Jun;20(12):1181-1194. doi: 10.1080/15384101.2021.1930357. Epub 2021 May 24. Cell Cycle. 2021. PMID: 34024251 Free PMC article.
-
Retracted Article: Circular RNA hsa_circ_0000467 modulates SGK1 to facilitate cell migration, metastasis, and EMT while repressing apoptosis in colorectal cancer by sponging miR-383-5p.RSC Adv. 2019 Nov 29;9(67):39294-39303. doi: 10.1039/c9ra07900a. eCollection 2019 Nov 27. RSC Adv. 2019. Retraction in: RSC Adv. 2021 Jan 20;11(7):4234. doi: 10.1039/d1ra90010b. PMID: 35540673 Free PMC article. Retracted.
-
Circular RNA hsa_circ_0007121 regulates proliferation, migration, invasion, and epithelial-mesenchymal transition of trophoblast cells by miR-182-5p/PGF axis in preeclampsia.Open Med (Wars). 2020 Oct 14;15(1):1061-1071. doi: 10.1515/med-2020-0230. eCollection 2020. Open Med (Wars). 2020. PMID: 33336062 Free PMC article.
-
Circ-0001801 contributes to cell proliferation, migration, invasion and epithelial to mesenchymal transition (EMT) in glioblastoma by regulating miR-628-5p/HMGB3 axis.Eur Rev Med Pharmacol Sci. 2019 Dec;23(24):10874-10885. doi: 10.26355/eurrev_201912_19791. Eur Rev Med Pharmacol Sci. 2019. PMID: 31858556
-
Downregulation of hsa_circ_0001681 suppresses epithelial-mesenchymal transition in thyroid carcinoma via targeting to miR-942-5p/TWIST1 signaling pathway.J Bioenerg Biomembr. 2021 Oct;53(5):609-620. doi: 10.1007/s10863-021-09907-2. Epub 2021 Jun 23. J Bioenerg Biomembr. 2021. PMID: 34164767
Cited by
-
Clinical implications of miR-195 in cancer: mechanisms, potential applications, and therapeutic strategies.J Cancer Res Clin Oncol. 2025 Apr 22;151(4):148. doi: 10.1007/s00432-025-06195-w. J Cancer Res Clin Oncol. 2025. PMID: 40261408 Free PMC article. Review.
-
The Role of the Dysregulation of circRNAs Expression in Glioblastoma Multiforme.J Mol Neurosci. 2025 Jan 22;75(1):9. doi: 10.1007/s12031-024-02285-5. J Mol Neurosci. 2025. PMID: 39841303 Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

