Computational design and engineering of self-assembling multivalent microproteins with therapeutic potential against SARS-CoV-2
- PMID: 38341574
- PMCID: PMC10858482
- DOI: 10.1186/s12951-024-02329-3
Computational design and engineering of self-assembling multivalent microproteins with therapeutic potential against SARS-CoV-2
Abstract
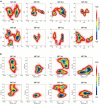
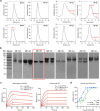
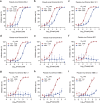
Multivalent drugs targeting homo-oligomeric viral surface proteins, such as the SARS-CoV-2 trimeric spike (S) protein, have the potential to elicit more potent and broad-spectrum therapeutic responses than monovalent drugs by synergistically engaging multiple binding sites on viral targets. However, rational design and engineering of nanoscale multivalent protein drugs are still lacking. Here, we developed a computational approach to engineer self-assembling trivalent microproteins that simultaneously bind to the three receptor binding domains (RBDs) of the S protein. This approach involves four steps: structure-guided linker design, molecular simulation evaluation of self-assembly, experimental validation of self-assembly state, and functional testing. Using this approach, we first designed trivalent constructs of the microprotein miniACE2 (MP) with different trimerization scaffolds and linkers, and found that one of the constructs (MP-5ff) showed high trimerization efficiency, good conformational homogeneity, and strong antiviral neutralizing activity. With its trimerization unit (5ff), we then engineered a trivalent nanobody (Tr67) that exhibited potent and broad neutralizing activity against the dominant Omicron variants, including XBB.1 and XBB.1.5. Cryo-EM complex structure confirmed that Tr67 stably binds to all three RBDs of the Omicron S protein in a synergistic form, locking them in the "3-RBD-up" conformation that could block human receptor (ACE2) binding and potentially facilitate immune clearance. Therefore, our approach provides an effective strategy for engineering potent protein drugs against SARS-CoV-2 and other deadly coronaviruses.
Keywords: Computational design; Cryo-EM; Microprotein; Nanobody; Protein therapeutics; SARS-CoV-2.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
Structure-guided design of a trivalent nanobody cluster targeting SARS-CoV-2 spike protein.Int J Biol Macromol. 2024 Jan;256(Pt 1):128191. doi: 10.1016/j.ijbiomac.2023.128191. Epub 2023 Nov 22. Int J Biol Macromol. 2024. PMID: 38000614
-
A mosaic-type trimeric RBD-based COVID-19 vaccine candidate induces potent neutralization against Omicron and other SARS-CoV-2 variants.Elife. 2022 Aug 25;11:e78633. doi: 10.7554/eLife.78633. Elife. 2022. PMID: 36004719 Free PMC article.
-
A Multivalent and Thermostable Nanobody Neutralizing SARS-CoV-2 Omicron (B.1.1.529).Int J Nanomedicine. 2023 Jan 19;18:353-367. doi: 10.2147/IJN.S387160. eCollection 2023. Int J Nanomedicine. 2023. PMID: 36700149 Free PMC article.
-
Structural and Computational Studies of the SARS-CoV-2 Spike Protein Binding Mechanisms with Nanobodies: From Structure and Dynamics to Avidity-Driven Nanobody Engineering.Int J Mol Sci. 2022 Mar 8;23(6):2928. doi: 10.3390/ijms23062928. Int J Mol Sci. 2022. PMID: 35328351 Free PMC article. Review.
-
Potent antibodies against immune invasive SARS-CoV-2 Omicron subvariants.Int J Biol Macromol. 2023 Sep 30;249:125997. doi: 10.1016/j.ijbiomac.2023.125997. Epub 2023 Jul 26. Int J Biol Macromol. 2023. PMID: 37499711 Review.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

