Molecular map of disulfidptosis-related genes in lung adenocarcinoma: the perspective toward immune microenvironment and prognosis
- PMID: 38342890
- PMCID: PMC10860275
- DOI: 10.1186/s13148-024-01632-y
Molecular map of disulfidptosis-related genes in lung adenocarcinoma: the perspective toward immune microenvironment and prognosis
Abstract
Background: Disulfidptosis is a recently discovered form of programmed cell death that could impact cancer development. Nevertheless, the prognostic significance of disulfidptosis-related genes (DRGs) in lung adenocarcinoma (LUAD) requires further clarification.
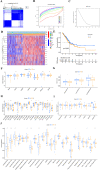
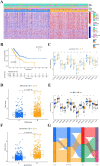
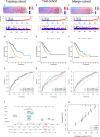
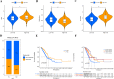
Methods: This study systematically explores the genetic and transcriptional variability, prognostic relevance, and expression profiles of DRGs. Clusters related to disulfidptosis were identified through consensus clustering. We used single-sample gene set enrichment analysis and ESTIMATE to assess the tumor microenvironment (TME) in different subgroups. We conducted a functional analysis of differentially expressed genes between subgroups, which involved gene ontology, the Kyoto encyclopedia of genes and genomes, and gene set variation analysis, in order to elucidate their functional status. Prognostic risk models were developed using univariate Cox regression and the least absolute shrinkage and selection operator regression. Additionally, single-cell clustering and cell communication analysis were conducted to enhance the understanding of the importance of signature genes. Lastly, qRT-PCR was employed to validate the prognostic model.
Results: Two clearly defined DRG clusters were identified through a consensus-based, unsupervised clustering analysis. Observations were made concerning the correlation between changes in multilayer DRG and various clinical characteristics, prognosis, and the infiltration of TME cells. A well-executed risk assessment model, known as the DRG score, was developed to predict the prognosis of LUAD patients. A high DRG score indicates increased TME cell infiltration, a higher mutation burden, elevated TME scores, and a poorer prognosis. Additionally, the DRG score showed a significant correlation with the tumor mutation burden score and the tumor immune dysfunction and exclusion score. Subsequently, a nomogram was established for facilitating the clinical application of the DRG score, showing good predictive ability and calibration. Additionally, crucial DRGs were further validated by single-cell sequencing data. Finally, crucial DRGs were further validated by qRT-PCR and immunohistochemistry.
Conclusion: Our new DRG signature risk score can predict the immune landscape and prognosis of LUAD. It also serves as a reference for LUAD's immunotherapy and chemotherapy.
Keywords: Disulfidptosis; Immune checkpoint inhibitors; Lung adenocarcinoma; Molecular subtypes; Tumor microenvironment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no potential competing interests.
Figures





Similar articles
-
Integrative analysis of single-cell and bulk RNA-sequencing data revealed disulfidptosis genes-based molecular subtypes and a prognostic signature in lung adenocarcinoma.Aging (Albany NY). 2024 Feb 5;16(3):2753-2773. doi: 10.18632/aging.205509. Epub 2024 Feb 5. Aging (Albany NY). 2024. PMID: 38319721 Free PMC article.
-
Toward precision oncology in LUAD: a prognostic model using single-cell sequencing and WGCNA based on a disulfidptosis relative gene signature.Front Immunol. 2025 May 21;16:1581915. doi: 10.3389/fimmu.2025.1581915. eCollection 2025. Front Immunol. 2025. PMID: 40469299 Free PMC article.
-
Bioinformatics-based analysis of the relationship between disulfidptosis and prognosis and treatment response in pancreatic cancer.Sci Rep. 2023 Dec 14;13(1):22218. doi: 10.1038/s41598-023-49752-4. Sci Rep. 2023. PMID: 38097783 Free PMC article. Clinical Trial.
-
Identification of a novel cuproptosis-related gene signature and integrative analyses in patients with lower-grade gliomas.Front Immunol. 2022 Aug 15;13:933973. doi: 10.3389/fimmu.2022.933973. eCollection 2022. Front Immunol. 2022. PMID: 36045691 Free PMC article. Review.
-
Development and experimental verification of a prognosis model for disulfidptosis-associated genes in HNSCC.Medicine (Baltimore). 2024 Mar 22;103(12):e37308. doi: 10.1097/MD.0000000000037308. Medicine (Baltimore). 2024. PMID: 38518012 Free PMC article. Review.
Cited by
-
Exploring the diagnostic and immune infiltration roles of disulfidptosis related genes in pulmonary hypertension.Respir Res. 2024 Oct 9;25(1):365. doi: 10.1186/s12931-024-02978-w. Respir Res. 2024. PMID: 39385167 Free PMC article.
-
Prognostic model of lung adenocarcinoma based on disulfidptosis-related genes and analysis of in vitro cell experiments for PPP1R14B in the model.Biol Direct. 2025 Jul 1;20(1):74. doi: 10.1186/s13062-025-00662-7. Biol Direct. 2025. PMID: 40598552 Free PMC article.
-
A Novel Disulfidptosis-Related Diagnostic Gene Signature and Differential Expression Validation in Ischaemic Cardiomyopathy.J Cell Mol Med. 2025 Mar;29(5):e70475. doi: 10.1111/jcmm.70475. J Cell Mol Med. 2025. PMID: 40070032 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

