Nutrient restriction-activated Fra-2 promotes tumor progression via IGF1R in miR-15a downmodulated pancreatic ductal adenocarcinoma
- PMID: 38342897
- PMCID: PMC10859382
- DOI: 10.1038/s41392-024-01740-4
Nutrient restriction-activated Fra-2 promotes tumor progression via IGF1R in miR-15a downmodulated pancreatic ductal adenocarcinoma
Abstract
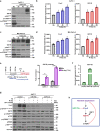
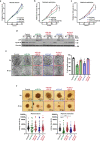
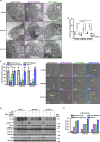
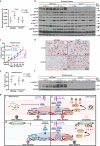
Pancreatic ductal adenocarcinoma (PDAC) is a lethal disease, characterized by an intense desmoplastic reaction that compresses blood vessels and limits nutrient supplies. PDAC aggressiveness largely relies on its extraordinary capability to thrive and progress in a challenging tumor microenvironment. Dysregulation of the onco-suppressor miR-15a has been extensively documented in PDAC. Here, we identified the transcription factor Fos-related antigen-2 (Fra-2) as a miR-15a target mediating the adaptive mechanism of PDAC to nutrient deprivation. We report that the IGF1 signaling pathway was enhanced in nutrient deprived PDAC cells and that Fra-2 and IGF1R were significantly overexpressed in miR-15a downmodulated PDAC patients. Mechanistically, we discovered that miR-15a repressed IGF1R expression via Fra-2 targeting. In miR-15a-low context, IGF1R hyperactivated mTOR, modulated the autophagic flux and sustained PDAC growth in nutrient deprivation. In a genetic mouse model, Mir15aKO PDAC showed Fra-2 and Igf1r upregulation and mTOR activation in response to diet restriction. Consistently, nutrient restriction improved the efficacy of IGF1R inhibition in a Fra-2 dependent manner. Overall, our results point to a crucial role of Fra-2 in the cellular stress response due to nutrient restriction typical of pancreatic cancer and support IGF1R as a promising and vulnerable target in miR-15a downmodulated PDAC.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests. C.M.C. is one of the Editors-in-Chief of Signal Transduction and Targeted Therapy, but he has not been involved in the process of the manuscript handling.
Figures


References
-
- Kalli M, Li R, Mills GB, Stylianopoulos T, Zervantonakis IK. Mechanical stress signaling in pancreatic cancer cells triggers p38 MAPK- and JNK-dependent cytoskeleton remodeling and promotes cell migration via Rac1/cdc42/Myosin II. Mol. Cancer Res. 2022;20:485–497. doi: 10.1158/1541-7786.MCR-21-0266. - DOI - PMC - PubMed
-
- Perera RM, Bardeesy N. Pancreatic cancer metabolism - breaking it down to build it back up. Cancer Discov. 2015;5:1247–1261. doi: 10.1158/2159-8290.CD-15-0671. - DOI - PMC - PubMed
-
- McAndrews KM, et al. Identification of functional heterogeneity of carcinoma-associated fibroblasts with distinct IL6-mediated therapy resistance in pancreatic cancer. Cancer Discov. 2022;12:1580–1597. doi: 10.1158/2159-8290.CD-20-1484. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

