Genomic vulnerability of a freshwater salmonid under climate change
- PMID: 38343776
- PMCID: PMC10853590
- DOI: 10.1111/eva.13602
Genomic vulnerability of a freshwater salmonid under climate change
Abstract
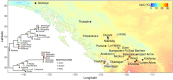
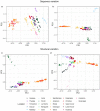
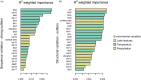
Understanding the adaptive potential of populations and species is pivotal for minimizing the loss of biodiversity in this era of rapid climate change. Adaptive potential has been estimated in various ways, including based on levels of standing genetic variation, presence of potentially beneficial alleles, and/or the severity of environmental change. Kokanee salmon, the non-migratory ecotype of sockeye salmon (Oncorhynchus nerka), is culturally and economically important and has already been impacted by the effects of climate change. To assess its climate vulnerability moving forward, we integrated analyses of standing genetic variation, genotype-environment associations, and climate modeling based on sequence and structural genomic variation from 224 whole genomes sampled from 22 lakes in British Columbia and Yukon (Canada). We found that variables for extreme temperatures, particularly warmer temperatures, had the most pervasive signature of selection in the genome and were the strongest predictors of levels of standing variation and of putatively adaptive genomic variation, both sequence and structural. Genomic offset estimates, a measure of climate vulnerability, were significantly correlated with higher increases in extreme warm temperatures, further highlighting the risk of summer heat waves that are predicted to increase in frequency in the future. Levels of standing genetic variation, an important metric for population viability and resilience, were not correlated with genomic offset. Nonetheless, our combined approach highlights the importance of integrating different sources of information and genomic data to formulate more comprehensive and accurate predictions on the vulnerability of populations and species to future climate change.
Keywords: Pacific salmon; climate vulnerability; local adaptation; standing variation; structural variants.
© 2023 The Authors. Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Abdul‐Aziz, O. I. , Mantua, N. J. , & Myers, K. W. (2011). Potential climate change impacts on thermal habitats of Pacific salmon (Oncorhynchus spp.) in the North Pacific Ocean and adjacent seas. Canadian Journal of Fisheries and Aquatic Sciences, 68(9), 1660–1680.
-
- Akopyan, M. , Tigano, A. , Jacobs, A. , Wilder, A. P. , Baumann, H. , & Therkildsen, N. O. (2022). Comparative linkage mapping uncovers recombination suppression across massive chromosomal inversions associated with local adaptation in Atlantic silversides. Molecular Ecology, 31(12), 3323–3341. - PubMed
-
- Andrews, K. R. , Seaborn, T. , Egan, J. P. , Fagnan, M. W. , New, D. D. , Chen, Z. , Hohenlohe, P. A. , Waits, L. P. , Caudill, C. C. , & Narum, S. R. (2023). Whole genome resequencing identifies local adaptation associated with environmental variation for redband trout. Molecular Ecology, 32(4), 800–818. - PMC - PubMed
-
- Barrett, R. D. H. , & Schluter, D. (2008). Adaptation from standing genetic variation. Trends in Ecology & Evolution, 23(1), 38–44. - PubMed
LinkOut - more resources
Full Text Sources

