This is a preprint.
Autoimmune diseases and risk of non-Hodgkin lymphoma: A Mendelian randomisation study
- PMID: 38343812
- PMCID: PMC10854352
- DOI: 10.1101/2024.01.20.24301459
Autoimmune diseases and risk of non-Hodgkin lymphoma: A Mendelian randomisation study
Update in
-
Autoimmune Diseases and Risk of Non-Hodgkin Lymphoma: A Mendelian Randomisation Study.Cancer Med. 2024 Nov;13(21):e70327. doi: 10.1002/cam4.70327. Cancer Med. 2024. PMID: 39506244 Free PMC article.
Abstract
Objective: To examine whether genetically predicted susceptibility to ten autoimmune diseases (Behçet's disease, coeliac disease, dermatitis herpetiformis, lupus, psoriasis, rheumatoid arthritis, sarcoidosis, Sjögren's syndrome, systemic sclerosis, and type 1 diabetes) is associated with risk of non-Hodgkin lymphoma (NHL).
Design: Two sample Mendelian randomization (MR) study.
Setting: Genome wide association studies (GWASs) of ten autoimmune diseases, NHL, and four NHL subtypes (i.e., follicular lymphoma, mature T/natural killer-cell lymphomas, non-follicular lymphoma, and other and unspecified types of NHL).
Analysis: We used data from the largest publicly available GWASs of European ancestry for each autoimmune disease, NHL, and NHL subtypes. For each autoimmune disease, we extracted single nucleotide polymorphisms (SNPs) strongly associated (P < 5×10-8) with that disease and that were independent of one another (R2 < 1×10-3) as genetic instruments. SNPs within the human leukocyte antigen region were not considered due to potential pleiotropy. Our primary MR analysis was the inverse-variance weighted analysis. Additionally, we conducted MR-Egger, weighted mode, and weighted median regression to address potential bias due to pleiotropy, and robust adjusted profile scores to address weak instrument bias. We carried out sensitivity analysis limited to the non-immune pathway for nominally significant findings. To account for multiple testing, we set the thresholds for statistical significance at P < 5×10-3.
Participants: The number of cases and controls identified in the relevant GWASs were 437 and 3,325 for Behçet's disease, 4,918 and 5,684 for coeliac disease, 435 and 341,188 for dermatitis herpetiformis, 4,576 and 8,039 for lupus, 11,988 and 275,335 for psoriasis, 22,350 and 74,823 for rheumatoid arthritis, 3,597 and 337,121 for sarcoidosis, 2,735 and 332,115 for Sjögren's syndrome, 9,095 and 17,584 for systemic sclerosis, 18,942 and 501,638 for type 1 diabetes, 2,400 and 410,350 for NHL; and 296 to 2,340 cases and 271,463 controls for NHL subtypes.
Exposures: Genetic variants predicting ten autoimmune diseases: Behçet's disease, coeliac disease, dermatitis herpetiformis, lupus, psoriasis, rheumatoid arthritis, sarcoidosis, Sjögren's syndrome, systemic sclerosis, and type 1 diabetes.
Main outcome measures: Estimated associations between genetically predicted susceptibility to ten autoimmune diseases and the risk of NHL.
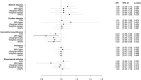
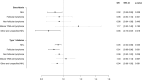
Results: The variance of each autoimmune disease explained by the SNPs ranged from 0.3% to 3.1%. Negative associations between type 1 diabetes and sarcoidosis and the risk of NHL were observed (odds ratio [OR] 0.95, 95% confidence interval [CI]: 0.92 to 0.98, P = 5×10-3, and OR 0.92, 95% CI: 0.85 to 0.99, P = 2.8×10-2, respectively). These findings were supported by the sensitivity analyses accounting for potential pleiotropy and weak instrument bias. No significant associations were found between the other eight autoimmune diseases and NHL risk. Of the NHL subtypes, type 1 diabetes was most strongly associated with follicular lymphoma (OR 0.91, 95% CI: 0.86 to 0.96, P = 1×10-3), while sarcoidosis was most strongly associated with other and unspecified NHL (OR 0.86, 95% CI: 0.75 to 0.97, P = 1.8×10-2).
Conclusions: These findings suggest that genetically predicted susceptibility to type 1 diabetes, and to some extent sarcoidosis, might reduce the risk of NHL. However, future studies with different datasets, approaches, and populations are warranted to further examine the potential associations between these autoimmune diseases and the risk of NHL.
Keywords: Autoimmune diseases; Mendelian randomisation; Non-Hodgkin lymphoma; Sarcoidosis; Type 1 diabetes.
Conflict of interest statement
Conflict of interest: All authors have completed the ICMJE uniform disclosure form at www.icmje.org/coi_disclosure.pdf and declare: In the past 36 months, Dr. Wallach reported receiving grant support from the FDA, Arnold Ventures, Johnson & Johnson through Yale University, and the National Institute on Alcohol Abuse and Alcoholism of the National Institutes of Health (NIH) under award 1K01AA028258; serving as a consultant for Hagens Berman Sobol Shapiro LLP and Dugan Law Firm APLC; and serving as a medRxiv affiliate. Dr. Ma received research funding from the NIH and the Frederick A. DeLuca Foundation and served as a consultant for Bristol Myers Squibb.
Figures


Similar articles
-
Autoimmune Diseases and Risk of Non-Hodgkin Lymphoma: A Mendelian Randomisation Study.Cancer Med. 2024 Nov;13(21):e70327. doi: 10.1002/cam4.70327. Cancer Med. 2024. PMID: 39506244 Free PMC article.
-
Exploring risk factors for autoimmune diseases complicated by non-hodgkin lymphoma through regulatory T cell immune-related traits: a Mendelian randomization study.Front Immunol. 2024 May 28;15:1374938. doi: 10.3389/fimmu.2024.1374938. eCollection 2024. Front Immunol. 2024. PMID: 38863695 Free PMC article.
-
[Genetic Causation Analysis of Hyperandrogenemia Testing Indicators and Preeclampsia].Sichuan Da Xue Xue Bao Yi Xue Ban. 2024 May 20;55(3):566-573. doi: 10.12182/20240560106. Sichuan Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 38948277 Free PMC article. Chinese.
-
The epidemiology of non-Hodgkin lymphoma.Pathology. 2005 Dec;37(6):409-19. doi: 10.1080/00313020500370192. Pathology. 2005. PMID: 16373224 Review.
-
Associations of non-Hodgkin Lymphoma (NHL) risk with autoimmune conditions according to putative NHL loci.Am J Epidemiol. 2015 Mar 15;181(6):406-21. doi: 10.1093/aje/kwu290. Epub 2015 Feb 23. Am J Epidemiol. 2015. PMID: 25713336 Free PMC article. Review.
Cited by
-
The causality between gut microbiota and non-Hodgkin lymphoma: a two-sample bidirectional Mendelian randomization study.Front Microbiol. 2024 May 27;15:1403825. doi: 10.3389/fmicb.2024.1403825. eCollection 2024. Front Microbiol. 2024. PMID: 38860220 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
