This is a preprint.
An Approach to Identifying Spatial Variability in Observed Infectious Disease Spread in a Prospective Time-Space Series with Applications to COVID-19 and Dengue Incidence
- PMID: 38343818
- PMCID: PMC10854290
- DOI: 10.21203/rs.3.rs-3859620/v1
An Approach to Identifying Spatial Variability in Observed Infectious Disease Spread in a Prospective Time-Space Series with Applications to COVID-19 and Dengue Incidence
Abstract
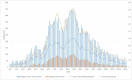
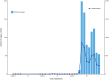
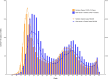
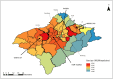
Most of the growing prospective analytic methods in space-time disease surveillance and intended functions of disease surveillance systems focus on earlier detection of disease outbreaks, disease clusters, or increased incidence. The spread of the virus such as SARS-CoV-2 has not been spatially and temporally uniform in an outbreak. With the identification of an infectious disease outbreak, recognizing and evaluating anomalies (excess and decline) of disease incidence spread at the time of occurrence during the course of an outbreak is a logical next step. We propose and formulate a hypergeometric probability model that investigates anomalies of infectious disease incidence spread at the time of occurrence in the timeline for many geographically described populations (e.g., hospitals, towns, counties) in an ongoing daily monitoring process. It is structured to determine whether the incidence grows or declines more rapidly in a region on the single current day or the most recent few days compared to the occurrence of the incidence during the previous few days relative to elsewhere in the surveillance period. The new method uses a time-varying baseline risk model, accounting for regularly (e.g., daily) updated information on disease incidence at the time of occurrence, and evaluates the probability of the deviation of particular frequencies to be attributed to sampling fluctuations, accounting for the unequal variances of the rates due to different population bases in geographical units. We attempt to present and illustrate a new model to advance the investigation of anomalies of infectious disease incidence spread by analyzing subsamples of spatiotemporal disease surveillance data from Taiwan on dengue and COVID-19 incidence which are mosquito-borne and contagious infectious diseases, respectively. Efficient R programs for computation are available to implement the two approximate formulae of the hypergeometric probability model for large numbers of events.
Figures
Similar articles
-
Assessing current temporal and space-time anomalies of disease incidence.PLoS One. 2017 Nov 13;12(11):e0188065. doi: 10.1371/journal.pone.0188065. eCollection 2017. PLoS One. 2017. PMID: 29131869 Free PMC article.
-
Effectiveness and cost-effectiveness of four different strategies for SARS-CoV-2 surveillance in the general population (CoV-Surv Study): a structured summary of a study protocol for a cluster-randomised, two-factorial controlled trial.Trials. 2021 Jan 8;22(1):39. doi: 10.1186/s13063-020-04982-z. Trials. 2021. PMID: 33419461 Free PMC article.
-
ID-Viewer: a visual analytics architecture for infectious diseases surveillance and response management in Pakistan.Public Health. 2016 May;134:72-85. doi: 10.1016/j.puhe.2016.01.006. Epub 2016 Feb 13. Public Health. 2016. PMID: 26880489
-
COVID-19 Variant Surveillance and Social Determinants in Central Massachusetts: Development Study.JMIR Form Res. 2022 Jun 13;6(6):e37858. doi: 10.2196/37858. JMIR Form Res. 2022. PMID: 35658093 Free PMC article.
-
Universal screening for SARS-CoV-2 infection: a rapid review.Cochrane Database Syst Rev. 2020 Sep 15;9(9):CD013718. doi: 10.1002/14651858.CD013718. Cochrane Database Syst Rev. 2020. PMID: 33502003 Free PMC article.
References
-
- Bühring W (1987) An analytic continuation of the hypergeometric series. Siam Journal on Mathematical Analysis 18: 884 – 889.
-
- Cliff AD, Ord JK (1981) Spatial Processes: Models & Applications. Taylor & Francis
-
- Cressie N (1992) Smoothing regional maps using empirical Bayes predictors. Geographical Analysis 24: 75–95.
-
- Cressie N, Chan NH (1989) Spatial Modeling of Regional Variables. Journal of the American Statistical Association 84: 393–401. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

