A Cross-section Metagenomics and 16S Ribosomal DNA Based Evaluation of the Bacterial and Archaeal Communities Resident in the Forumad Chromite Mine, Northeastern of Iran
- PMID: 38344319
- PMCID: PMC10858365
- DOI: 10.30498/ijb.2022.240607.2818
A Cross-section Metagenomics and 16S Ribosomal DNA Based Evaluation of the Bacterial and Archaeal Communities Resident in the Forumad Chromite Mine, Northeastern of Iran
Abstract
Background: The Forumad chromite area from Sabzevar ophiolite belt, Northeastern Iran, is an environment with high concentration of heavy metals, particularly chromite and magnesite minerals, containing chromium and magnesium.
Objectives: In this study for the first time, we analyzed and report the diversity of microbial (bacterial and archaeal) community inhabiting in Forumad chromite mine environment using metagenomics approach.
Materials and methods: Samples were obtained from different areas of the mine, and total DNA was extracted from water and soil samples. 16S rDNA was amplified using universal primers and the PCR products were cloned in pTz57R/T plasmid. Then, 43% of the positive clones were randomly sequenced. BLAST program in NCBI and EzTaxon databases were used to identify similar 16S rDNA sequences. Phylogenetic analysis was performed using the MEGA5 software and multiple alignments of sequences.
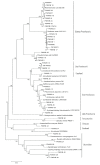
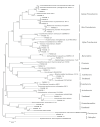
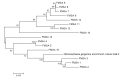
Results: In the phylogenetic analyses, proteobacteria, which contains many heavy metals tolerant bacteria especially chromium, were the dominant population in bacterial libraries with Rheinheimera and Cedecaeas the most abundant genuses. Other phyla were Bacteroidetes, Firmicutes, Verrucomicrobia, Chloroflexi, Actinobacteria, Acidobacteria, Cyanobacteria, Gemmatimonadetes, and Planctomycetes. In the archaeal clone library, all the sequences were related to the phylum Thaumarchaeota. Further, 68.6% of the sequences had less than 98.7℅ similarity with the recorded strains which could represent new taxons.
Conclusions: The results showed that there was a high microbial diversity in the Forumad chromite area. These results can be used for detoxification and bioremediation of regions contaminated with heavy metals, although more studies are needed.
Keywords: 16S rRNA; Forumad chromite mine; Metagenomics; Microbial diversity.
Copyright: © 2021 The Author(s); Published by Iranian Journal of Biotechnology.
Conflict of interest statement
The authors confirm that this article content has no conflicts of interest.
Figures
Similar articles
-
Analysis of microbial communities in heavy metals-contaminated soils using the metagenomic approach.Ecotoxicology. 2018 Nov;27(9):1281-1291. doi: 10.1007/s10646-018-1981-x. Epub 2018 Sep 21. Ecotoxicology. 2018. PMID: 30242595
-
[Prokaryotic microbial diversity of the ancient salt deposits in the Kunming Salt Mine, P.R. China].Wei Sheng Wu Xue Bao. 2007 Apr;47(2):295-300. Wei Sheng Wu Xue Bao. 2007. PMID: 17552238 Chinese.
-
Delineating bacterial community structure of polluted soil samples collected from cancer prone belt of Punjab, India.3 Biotech. 2015 Oct;5(5):727-734. doi: 10.1007/s13205-014-0270-5. Epub 2015 Jan 7. 3 Biotech. 2015. PMID: 28324527 Free PMC article.
-
Molecular analyses of microbial diversity associated with the Lonar soda lake in India: an impact crater in a basalt area.Res Microbiol. 2006 Dec;157(10):928-37. doi: 10.1016/j.resmic.2006.08.005. Epub 2006 Oct 9. Res Microbiol. 2006. PMID: 17070674
-
Molecular analysis of prokaryotic diversity in the deep subsurface of the former Homestake gold mine, South Dakota, USA.J Microbiol. 2009 Aug;47(4):371-84. doi: 10.1007/s12275-008-0249-1. Epub 2009 Sep 9. J Microbiol. 2009. PMID: 19763410
References
-
- Streit WR, Daniel R. Metagenomics: Methods and Protocols, Methods in Molecular Biology. Springer Science+Business Media. 2010;668:chapter 1. doi: 10.1007/978-1-4939-6691-2. - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
