Deciphering the intricate hierarchical gene regulatory network: unraveling multi-level regulation and modifications driving secondary cell wall formation
- PMID: 38344650
- PMCID: PMC10857936
- DOI: 10.1093/hr/uhad281
Deciphering the intricate hierarchical gene regulatory network: unraveling multi-level regulation and modifications driving secondary cell wall formation
Abstract
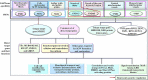
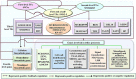
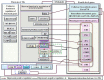
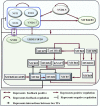
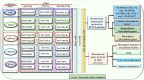
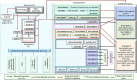
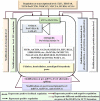
Wood quality is predominantly determined by the amount and the composition of secondary cell walls (SCWs). Consequently, unraveling the molecular regulatory mechanisms governing SCW formation is of paramount importance for genetic engineering aimed at enhancing wood properties. Although SCW formation is known to be governed by a hierarchical gene regulatory network (HGRN), our understanding of how a HGRN operates and regulates the formation of heterogeneous SCWs for plant development and adaption to ever-changing environment remains limited. In this review, we examined the HGRNs governing SCW formation and highlighted the significant key differences between herbaceous Arabidopsis and woody plant poplar. We clarified many confusions in existing literatures regarding the HGRNs and their orthologous gene names and functions. Additionally, we revealed many network motifs including feed-forward loops, feed-back loops, and negative and positive autoregulation in the HGRNs. We also conducted a thorough review of post-transcriptional and post-translational aspects, protein-protein interactions, and epigenetic modifications of the HGRNs. Furthermore, we summarized how the HGRNs respond to environmental factors and cues, influencing SCW biosynthesis through regulatory cascades, including many regulatory chains, wiring regulations, and network motifs. Finally, we highlighted the future research directions for gaining a further understanding of molecular regulatory mechanisms underlying SCW formation.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nanjing Agricultural University.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Tissue and cell-specific transcriptomes in cotton reveal the subtleties of gene regulation underlying the diversity of plant secondary cell walls.BMC Genomics. 2017 Jul 18;18(1):539. doi: 10.1186/s12864-017-3902-4. BMC Genomics. 2017. PMID: 28720072 Free PMC article.
-
Hormonal regulation of secondary cell wall formation.J Exp Bot. 2015 Aug;66(16):5015-27. doi: 10.1093/jxb/erv222. Epub 2015 May 22. J Exp Bot. 2015. PMID: 26002972 Review.
-
Comparative genomic analysis of the R2R3 MYB secondary cell wall regulators of Arabidopsis, poplar, rice, maize, and switchgrass.BMC Plant Biol. 2014 May 18;14:135. doi: 10.1186/1471-2229-14-135. BMC Plant Biol. 2014. PMID: 24885077 Free PMC article.
-
Blue Light Regulates Secondary Cell Wall Thickening via MYC2/MYC4 Activation of the NST1-Directed Transcriptional Network in Arabidopsis.Plant Cell. 2018 Oct;30(10):2512-2528. doi: 10.1105/tpc.18.00315. Epub 2018 Sep 21. Plant Cell. 2018. PMID: 30242037 Free PMC article.
-
NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants.Front Plant Sci. 2015 May 5;6:288. doi: 10.3389/fpls.2015.00288. eCollection 2015. Front Plant Sci. 2015. PMID: 25999964 Free PMC article. Review.
Cited by
-
Genome-Wide Identification and Expression Profiling of Velvet Complex Transcription Factors in Populus alba × Populus glandulosa.Int J Mol Sci. 2024 Mar 31;25(7):3926. doi: 10.3390/ijms25073926. Int J Mol Sci. 2024. PMID: 38612736 Free PMC article.
-
Genome-Wide Characterization and Expression Profile of the Jumonji-C Family Genes in Populus alba × Populus glandulosa Reveal Their Potential Roles in Wood Formation.Int J Mol Sci. 2025 Jun 13;26(12):5666. doi: 10.3390/ijms26125666. Int J Mol Sci. 2025. PMID: 40565130 Free PMC article.
-
Computational Reconstruction of the Transcription Factor Regulatory Network Induced by Auxin in Arabidopsis thaliana L.Plants (Basel). 2024 Jul 10;13(14):1905. doi: 10.3390/plants13141905. Plants (Basel). 2024. PMID: 39065433 Free PMC article.
-
CcNAC6 Acts as a Positive Regulator of Secondary Cell Wall Synthesis in Sudan Grass (Sorghum sudanense S.).Plants (Basel). 2024 May 14;13(10):1352. doi: 10.3390/plants13101352. Plants (Basel). 2024. PMID: 38794423 Free PMC article.
-
Transcription factor PagMYB31 positively regulates cambium activity and negatively regulates xylem development in poplar.Plant Cell. 2024 May 1;36(5):1806-1828. doi: 10.1093/plcell/koae040. Plant Cell. 2024. PMID: 38339982 Free PMC article.
References
-
- Ruzicka K, Ursache R, Hejatko J. et al. . Xylem development - from the cradle to the grave. New Phytol. 2015;207:519–35 - PubMed
-
- Anthony B, Serra S, Musacchi S. Optimization of light interception, leaf area and yield in “WA38”: comparisons among training systems, rootstocks and pruning techniques. Agronomy. 2020;10:689
-
- Read PE, Bavougian CM. Woody ornamentals. In: Dixon GR, Aldous DE (eds.), Horticulture: Plants for People and Places, Volume 2: Environmental Horticulture. New York: Springer, 2014, 619–44
-
- Peschiutta ML, Bucci SJ, Scholz FG. et al. . Leaf and stem hydraulic traits in relation to growth, water use and fruit yield in Prunus avium L. cultivars. Trees-Structure and Function. 2013;27:1559–69
LinkOut - more resources
Full Text Sources

