A natural variation in OsDSK2a modulates plant growth and salt tolerance through phosphorylation by SnRK1A in rice
- PMID: 38346083
- PMCID: PMC11182596
- DOI: 10.1111/pbi.14308
A natural variation in OsDSK2a modulates plant growth and salt tolerance through phosphorylation by SnRK1A in rice
Abstract
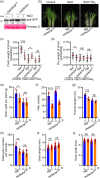
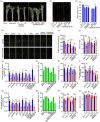
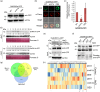
Plants grow rapidly for maximal production under optimal conditions; however, they adopt a slower growth strategy to maintain survival when facing environmental stresses. As salt stress restricts crop architecture and grain yield, identifying genetic variations associated with growth and yield responses to salinity is critical for breeding optimal crop varieties. OsDSK2a is a pivotal modulator of plant growth and salt tolerance via the modulation of gibberellic acid (GA) metabolism; however, its regulation remains unclear. Here, we showed that OsDSK2a can be phosphorylated at the second amino acid (S2) to maintain its stability. The gene-edited mutant osdsk2aS2G showed decreased plant height and enhanced salt tolerance. SnRK1A modulated OsDSK2a-S2 phosphorylation and played a substantial role in GA metabolism. Genetic analysis indicated that SnRK1A functions upstream of OsDSK2a and affects plant growth and salt tolerance. Moreover, SnRK1A activity was suppressed under salt stress, resulting in decreased phosphorylation and abundance of OsDSK2a. Thus, SnRK1A preserves the stability of OsDSK2a to maintain plant growth under normal conditions, and reduces the abundance of OsDSK2a to limit growth under salt stress. Haplotype analysis using 3 K-RG data identified a natural variation in OsDSK2a-S2. The allele of OsDSK2a-G downregulates plant height and improves salt-inhibited grain yield. Thus, our findings revealed a new mechanism for OsDSK2a stability and provided a valuable target for crop breeding to overcome yield limitations under salinity stress.
Keywords: gibberellic acid metabolism; natural variation; phosphorylation; plant growth; salt tolerance.
© 2024 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests of this work.
Figures




Similar articles
-
The Ubiquitin-Binding Protein OsDSK2a Mediates Seedling Growth and Salt Responses by Regulating Gibberellin Metabolism in Rice.Plant Cell. 2020 Feb;32(2):414-428. doi: 10.1105/tpc.19.00593. Epub 2019 Dec 11. Plant Cell. 2020. PMID: 31826965 Free PMC article.
-
miR396b/GRF6 module contributes to salt tolerance in rice.Plant Biotechnol J. 2024 Aug;22(8):2079-2092. doi: 10.1111/pbi.14326. Epub 2024 Mar 7. Plant Biotechnol J. 2024. PMID: 38454780 Free PMC article.
-
SnRK1A-interacting negative regulators modulate the nutrient starvation signaling sensor SnRK1 in source-sink communication in cereal seedlings under abiotic stress.Plant Cell. 2014 Feb;26(2):808-27. doi: 10.1105/tpc.113.121939. Epub 2014 Feb 25. Plant Cell. 2014. PMID: 24569770 Free PMC article.
-
GROWTH REGULATING FACTOR 7-mediated arbutin metabolism enhances rice salt tolerance.Plant Cell. 2024 Jul 31;36(8):2834-2850. doi: 10.1093/plcell/koae140. Plant Cell. 2024. PMID: 38701348 Free PMC article.
-
Root Development and Stress Tolerance in rice: The Key to Improving Stress Tolerance without Yield Penalties.Int J Mol Sci. 2020 Mar 6;21(5):1807. doi: 10.3390/ijms21051807. Int J Mol Sci. 2020. PMID: 32155710 Free PMC article. Review.
Cited by
-
The Regulation of ROS and Phytohormones in Balancing Crop Yield and Salt Tolerance.Antioxidants (Basel). 2025 Jan 7;14(1):63. doi: 10.3390/antiox14010063. Antioxidants (Basel). 2025. PMID: 39857397 Free PMC article. Review.
-
PAM-relaxed and temperature-tolerant CRISPR-Mb3Cas12a single transcript unit systems for efficient singular and multiplexed genome editing in rice, maize, and tomato.Plant Biotechnol J. 2025 Jan;23(1):156-173. doi: 10.1111/pbi.14486. Epub 2024 Oct 10. Plant Biotechnol J. 2025. PMID: 39387219 Free PMC article.
References
-
- Baena‐Gonzalez, E. and Hanson, J. (2017) Shaping plant development through the SnRK1‐TOR metabolic regulators. Curr. Opin. Plant Biol. 35, 152–157. - PubMed
-
- Baena‐Gonzalez, E. and Lunn, J.E. (2020) SnRK1 and trehalose 6‐phosphate – two ancient pathways converge to regulate plant metabolism and growth. Curr. Opin. Plant Biol. 55, 52–59. - PubMed
-
- Baena‐Gonzalez, E. , Rolland, F. , Thevelein, J.M. and Sheen, J. (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature, 448, 938–942. - PubMed
MeSH terms
Substances
Grants and funding
- 2022YFD1201700/National Key Research and Development Program of China
- CAAS-ZDRW202201/Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences
- 32030079/National Natural Science Foundation of China
- 32171949/National Natural Science Foundation of China
- 1610392020004/Fundamental Research Funds for Central Non-profit Scientific Institution
LinkOut - more resources
Full Text Sources
Research Materials

