A methylation-phosphorylation switch controls EZH2 stability and hematopoiesis
- PMID: 38346162
- PMCID: PMC10901513
- DOI: 10.7554/eLife.86168
A methylation-phosphorylation switch controls EZH2 stability and hematopoiesis
Abstract
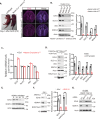
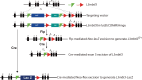
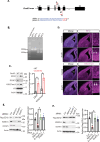
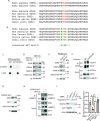
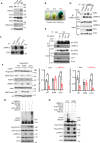
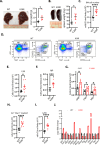
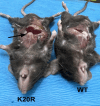
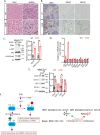
The Polycomb Repressive Complex 2 (PRC2) methylates H3K27 to regulate development and cell fate by transcriptional silencing. Alteration of PRC2 is associated with various cancers. Here, we show that mouse Kdm1a deletion causes a dramatic reduction of PRC2 proteins, whereas mouse null mutation of L3mbtl3 or Dcaf5 results in PRC2 accumulation and increased H3K27 trimethylation. The catalytic subunit of PRC2, EZH2, is methylated at lysine 20 (K20), promoting EZH2 proteolysis by L3MBTL3 and the CLR4DCAF5 ubiquitin ligase. KDM1A (LSD1) demethylates the methylated K20 to stabilize EZH2. K20 methylation is inhibited by AKT-mediated phosphorylation of serine 21 in EZH2. Mouse Ezh2K20R/K20R mutants develop hepatosplenomegaly associated with high GFI1B expression, and Ezh2K20R/K20R mutant bone marrows expand hematopoietic stem cells and downstream hematopoietic populations. Our studies reveal that EZH2 is regulated by methylation-dependent proteolysis, which is negatively controlled by AKT-mediated S21 phosphorylation to establish a methylation-phosphorylation switch to regulate the PRC2 activity and hematopoiesis.
Keywords: AKT; CRL4; DCAF5; EZH2; KDM1A; L3MBTL3; PRC2; SET7; biochemistry; cancer biology; chemical biology; mouse.
© 2024, Guo et al.
Conflict of interest statement
PG, RL, KR, TT, HS, HZ No competing interests declared
Figures















Update of
- doi: 10.1101/2023.02.02.526767
Similar articles
-
Distinct Stimulatory Mechanisms Regulate the Catalytic Activity of Polycomb Repressive Complex 2.Mol Cell. 2018 May 3;70(3):435-448.e5. doi: 10.1016/j.molcel.2018.03.019. Epub 2018 Apr 19. Mol Cell. 2018. PMID: 29681498 Free PMC article.
-
EZH2 variants differentially regulate polycomb repressive complex 2 in histone methylation and cell differentiation.Epigenetics Chromatin. 2018 Dec 6;11(1):71. doi: 10.1186/s13072-018-0242-9. Epigenetics Chromatin. 2018. PMID: 30522506 Free PMC article.
-
Regulation of histone methylation by automethylation of PRC2.Genes Dev. 2019 Oct 1;33(19-20):1416-1427. doi: 10.1101/gad.328849.119. Epub 2019 Sep 5. Genes Dev. 2019. PMID: 31488576 Free PMC article.
-
The role of EZH1 and EZH2 in development and cancer.BMB Rep. 2022 Dec;55(12):595-601. doi: 10.5483/BMBRep.2022.55.12.174. BMB Rep. 2022. PMID: 36476271 Free PMC article. Review.
-
The roles of EZH2 in cancer and its inhibitors.Med Oncol. 2023 May 6;40(6):167. doi: 10.1007/s12032-023-02025-6. Med Oncol. 2023. PMID: 37148376 Free PMC article. Review.
Cited by
-
Dynamic In Vivo Mapping of the Methylproteome Using a Chemoenzymatic Approach.J Am Chem Soc. 2025 Mar 5;147(9):7214-7230. doi: 10.1021/jacs.4c08175. Epub 2025 Feb 25. J Am Chem Soc. 2025. PMID: 39996454 Free PMC article.
-
Identification of DCAF5 as a novel cancer prognostic and immunotherapy biomarker through pan-cancer analysis and renal clear cell carcinoma clinical data validation.Discov Oncol. 2025 Jun 16;16(1):1119. doi: 10.1007/s12672-025-02974-6. Discov Oncol. 2025. PMID: 40522583 Free PMC article.
-
Epigenetic control and manipulation of neuronal maturation timing.Curr Opin Genet Dev. 2024 Apr;85:102164. doi: 10.1016/j.gde.2024.102164. Epub 2024 Feb 27. Curr Opin Genet Dev. 2024. PMID: 38412562 Free PMC article. Review.
References
-
- Abd Al Kader L, Oka T, Takata K, Sun X, Sato H, Murakami I, Toji T, Manabe A, Kimura H, Yoshino T. In aggressive variants of non-hodgkin lymphomas, Ezh2 is strongly expressed and polycomb repressive complex PRC1.4 dominates over PRC1.2. Virchows Archiv. 2013;463:697–711. doi: 10.1007/s00428-013-1428-y. - DOI - PubMed
-
- Adolfsson J, Borge OJ, Bryder D, Theilgaard-Mönch K, Astrand-Grundström I, Sitnicka E, Sasaki Y, Jacobsen SE. Upregulation of Flt3 expression within the bone marrow Lin(-)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15:659–669. doi: 10.1016/s1074-7613(01)00220-5. - DOI - PubMed
-
- Andlauer TFM, Buck D, Antony G, Bayas A, Bechmann L, Berthele A, Chan A, Gasperi C, Gold R, Graetz C, Haas J, Hecker M, Infante-Duarte C, Knop M, Kümpfel T, Limmroth V, Linker RA, Loleit V, Luessi F, Meuth SG, Mühlau M, Nischwitz S, Paul F, Pütz M, Ruck T, Salmen A, Stangel M, Stellmann JP, Stürner KH, Tackenberg B, Then Bergh F, Tumani H, Warnke C, Weber F, Wiendl H, Wildemann B, Zettl UK, Ziemann U, Zipp F, Arloth J, Weber P, Radivojkov-Blagojevic M, Scheinhardt MO, Dankowski T, Bettecken T, Lichtner P, Czamara D, Carrillo-Roa T, Binder EB, Berger K, Bertram L, Franke A, Gieger C, Herms S, Homuth G, Ising M, Jöckel KH, Kacprowski T, Kloiber S, Laudes M, Lieb W, Lill CM, Lucae S, Meitinger T, Moebus S, Müller-Nurasyid M, Nöthen MM, Petersmann A, Rawal R, Schminke U, Strauch K, Völzke H, Waldenberger M, Wellmann J, Porcu E, Mulas A, Pitzalis M, Sidore C, Zara I, Cucca F, Zoledziewska M, Ziegler A, Hemmer B, Müller-Myhsok B. Novel multiple sclerosis susceptibility loci implicated in epigenetic regulation. Science Advances. 2016;2:e1501678. doi: 10.1126/sciadv.1501678. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

