Applying valency-based immuno-selection to generate broadly cross-reactive antibodies against influenza hemagglutinins
- PMID: 38346952
- PMCID: PMC10861589
- DOI: 10.1038/s41467-024-44889-w
Applying valency-based immuno-selection to generate broadly cross-reactive antibodies against influenza hemagglutinins
Abstract
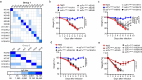
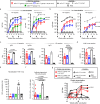
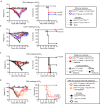
Conserved epitopes shared between virus subtypes are often subdominant, making it difficult to induce broadly reactive antibodies by immunization. Here, we generate a plasmid DNA mix vaccine that encodes protein heterodimers with sixteen different influenza A virus hemagglutinins (HA) representing all HA subtypes except H1 (group 1) and H7 (group 2). Each single heterodimer expresses two different HA subtypes and is targeted to MHC class II on antigen presenting cells (APC). Female mice immunized with the plasmid mix produce antibodies not only against the 16 HA subtypes, but also against non-included H1 and H7. We demonstrate that individual antibody molecules cross-react between different HAs. Furthermore, the mix vaccine induces T cell responses to conserved HA epitopes. Immunized mice are partially protected against H1 viruses. The results show that application of valency-based immuno-selection to diversified antigens can be used to direct antibody responses towards conserved (subdominant) epitopes on viral antigens.
© 2024. The Author(s).
Conflict of interest statement
The authors declare the following competing interest: the research funding did not have any role in the conceptualization, design, data collection, analysis, decision to publish, or preparation of the manuscript. B.B. holds shares in Nykode A.S. The TTO office of the University of Oslo and Oslo University Hospital, Inven2, has filed a patent application, WO 2019048928 National phase, “Vaccine molecules”, related to the valency-based immune-selection strategy described in the manuscript. R.B. and B.B. are inventors of this patent. The remaining authors declare no competing interests.
Figures





Similar articles
-
Protection against H1N1 influenza challenge by a DNA vaccine expressing H3/H1 subtype hemagglutinin combined with MHC class II-restricted epitopes.Virol J. 2010 Dec 7;7:363. doi: 10.1186/1743-422X-7-363. Virol J. 2010. PMID: 21134292 Free PMC article.
-
Induction of broadly reactive anti-hemagglutinin stalk antibodies by an H5N1 vaccine in humans.J Virol. 2014 Nov;88(22):13260-8. doi: 10.1128/JVI.02133-14. Epub 2014 Sep 10. J Virol. 2014. PMID: 25210189 Free PMC article.
-
Broadly neutralizing antibodies to combat influenza virus infection.Antiviral Res. 2024 Jan;221:105785. doi: 10.1016/j.antiviral.2023.105785. Epub 2023 Dec 23. Antiviral Res. 2024. PMID: 38145757 Review.
-
Broadly Protective CD8+ T Cell Immunity to Highly Conserved Epitopes Elicited by Heat Shock Protein gp96-Adjuvanted Influenza Monovalent Split Vaccine.J Virol. 2021 May 24;95(12):e00507-21. doi: 10.1128/JVI.00507-21. Print 2021 May 24. J Virol. 2021. PMID: 33827939 Free PMC article.
-
Hide and seek: interplay between influenza viruses and B cells.Int Immunol. 2020 Sep 8;32(9):605-611. doi: 10.1093/intimm/dxaa028. Int Immunol. 2020. PMID: 32304215 Free PMC article. Review.
References
-
- WHO. Influenza (Seasonal) Fact Sheet 2023. Last update: January 12, 2023. Accessed 6 August 2023. https://www.who.int/news-room/fact-sheets/detail/influenza -(seasonal).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

