Chromatin attachment to the nuclear matrix represses hypocotyl elongation in Arabidopsis thaliana
- PMID: 38346986
- PMCID: PMC10861482
- DOI: 10.1038/s41467-024-45577-5
Chromatin attachment to the nuclear matrix represses hypocotyl elongation in Arabidopsis thaliana
Abstract
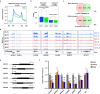
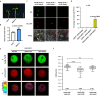
The nuclear matrix is a nuclear compartment that has diverse functions in chromatin regulation and transcription. However, how this structure influences epigenetic modifications and gene expression in plants is largely unknown. In this study, we show that a nuclear matrix binding protein, AHL22, together with the two transcriptional repressors FRS7 and FRS12, regulates hypocotyl elongation by suppressing the expression of a group of genes known as SMALL AUXIN UP RNAs (SAURs) in Arabidopsis thaliana. The transcriptional repression of SAURs depends on their attachment to the nuclear matrix. The AHL22 complex not only brings these SAURs, which contain matrix attachment regions (MARs), to the nuclear matrix, but it also recruits the histone deacetylase HDA15 to the SAUR loci. This leads to the removal of H3 acetylation at the SAUR loci and the suppression of hypocotyl elongation. Taken together, our results indicate that MAR-binding proteins act as a hub for chromatin and epigenetic regulators. Moreover, we present a mechanism by which nuclear matrix attachment to chromatin regulates histone modifications, transcription, and hypocotyl elongation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

