The metabolism-related lncRNA signature predicts the prognosis of breast cancer patients
- PMID: 38347041
- PMCID: PMC10861477
- DOI: 10.1038/s41598-024-53716-7
The metabolism-related lncRNA signature predicts the prognosis of breast cancer patients
Abstract
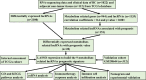
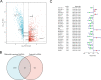
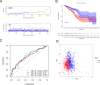
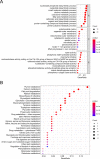
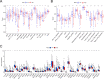
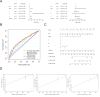
Long non-coding RNAs (lncRNAs) involved in metabolism are recognized as significant factors in breast cancer (BC) progression. We constructed a novel prognostic signature for BC using metabolism-related lncRNAs and investigated their underlying mechanisms. The training and validation cohorts were established from BC patients acquired from two public sources: The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO). The prognostic signature of metabolism-related lncRNAs was constructed using the least absolute shrinkage and selection operator (LASSO) cox regression analysis. We developed and validated a new prognostic risk model for BC using the signature of metabolism-related lncRNAs (SIRLNT, SIAH2-AS1, MIR205HG, USP30-AS1, MIR200CHG, TFAP2A-AS1, AP005131.2, AL031316.1, C6orf99). The risk score obtained from this signature was proven to be an independent prognostic factor for BC patients, resulting in a poor overall survival (OS) for individuals in the high-risk group. The area under the curve (AUC) for OS at three and five years were 0.67 and 0.65 in the TCGA cohort, and 0.697 and 0.68 in the GEO validation cohort, respectively. The prognostic signature demonstrated a robust association with the immunological state of BC patients. Conventional chemotherapeutics, such as docetaxel and paclitaxel, showed greater efficacy in BC patients classified as high-risk. A nomogram with a c-index of 0.764 was developed to forecast the survival time of BC patients, considering their risk score and age. The silencing of C6orf99 markedly decreased the proliferation, migration, and invasion capacities in MCF-7 cells. Our study identified a signature of metabolism-related lncRNAs that predicts outcomes in BC patients and could assist in tailoring personalized prevention and treatment plans.
Keywords: Bioinformatics; Breast cancer; Metabolism-related lncRNAs; Prediction signature; Risk score.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

