Single-nucleoid architecture reveals heterogeneous packaging of mitochondrial DNA
- PMID: 38347148
- PMCID: PMC11370055
- DOI: 10.1038/s41594-024-01225-6
Single-nucleoid architecture reveals heterogeneous packaging of mitochondrial DNA
Abstract
Cellular metabolism relies on the regulation and maintenance of mitochondrial DNA (mtDNA). Hundreds to thousands of copies of mtDNA exist in each cell, yet because mitochondria lack histones or other machinery important for nuclear genome compaction, it remains unresolved how mtDNA is packaged into individual nucleoids. In this study, we used long-read single-molecule accessibility mapping to measure the compaction of individual full-length mtDNA molecules at near single-nucleotide resolution. We found that, unlike the nuclear genome, human mtDNA largely undergoes all-or-none global compaction, with most nucleoids existing in an inaccessible, inactive state. Highly accessible mitochondrial nucleoids are co-occupied by transcription and replication components and selectively form a triple-stranded displacement loop structure. In addition, we showed that the primary nucleoid-associated protein TFAM directly modulates the fraction of inaccessible nucleoids both in vivo and in vitro, acting consistently with a nucleation-and-spreading mechanism to coat and compact mitochondrial nucleoids. Together, these findings reveal the primary architecture of mtDNA packaging and regulation in human cells.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests
The authors declare no competing interests.
Figures
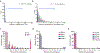




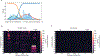








Similar articles
-
The TFAM-to-mtDNA ratio defines inner-cellular nucleoid populations with distinct activity levels.Cell Rep. 2021 Nov 23;37(8):110000. doi: 10.1016/j.celrep.2021.110000. Cell Rep. 2021. PMID: 34818548
-
Organization of DNA in Mammalian Mitochondria.Int J Mol Sci. 2019 Jun 5;20(11):2770. doi: 10.3390/ijms20112770. Int J Mol Sci. 2019. PMID: 31195723 Free PMC article. Review.
-
Phase separation drives the self-assembly of mitochondrial nucleoids for transcriptional modulation.Nat Struct Mol Biol. 2021 Nov;28(11):900-908. doi: 10.1038/s41594-021-00671-w. Epub 2021 Oct 28. Nat Struct Mol Biol. 2021. PMID: 34711968
-
Live imaging reveals the dynamics and regulation of mitochondrial nucleoids during the cell cycle in Fucci2-HeLa cells.Sci Rep. 2017 Sep 12;7(1):11257. doi: 10.1038/s41598-017-10843-8. Sci Rep. 2017. PMID: 28900194 Free PMC article.
-
mtDNA makes a U-turn for the mitochondrial nucleoid.Trends Cell Biol. 2013 Sep;23(9):457-63. doi: 10.1016/j.tcb.2013.04.009. Epub 2013 May 27. Trends Cell Biol. 2013. PMID: 23721879 Review.
Cited by
-
A kinetic dichotomy between mitochondrial and nuclear gene expression processes.Mol Cell. 2024 Apr 18;84(8):1541-1555.e11. doi: 10.1016/j.molcel.2024.02.028. Epub 2024 Mar 18. Mol Cell. 2024. PMID: 38503286 Free PMC article.
-
Profiling the epigenome using long-read sequencing.Nat Genet. 2025 Jan;57(1):27-41. doi: 10.1038/s41588-024-02038-5. Epub 2025 Jan 8. Nat Genet. 2025. PMID: 39779955 Review.
-
Mitochondrial DNA leakage: underlying mechanisms and therapeutic implications in neurological disorders.J Neuroinflammation. 2025 Feb 7;22(1):34. doi: 10.1186/s12974-025-03363-0. J Neuroinflammation. 2025. PMID: 39920753 Free PMC article. Review.
-
Opportunities and challenges of single-cell and spatially resolved genomics methods for neuroscience discovery.Nat Neurosci. 2024 Dec;27(12):2292-2309. doi: 10.1038/s41593-024-01806-0. Epub 2024 Dec 3. Nat Neurosci. 2024. PMID: 39627587 Free PMC article. Review.
-
Age-induced changes in skeletal muscle mitochondrial DNA synthesis, quantity, and quality in genetically unique rats.Geroscience. 2025 Feb;47(1):851-862. doi: 10.1007/s11357-024-01344-4. Epub 2024 Sep 23. Geroscience. 2025. PMID: 39312152 Free PMC article.
References
-
- Ojala D, Crews S, Montoya J, Gelfand R & Attardi G A small polyadenylated RNA (7 S RNA), containing a putative ribosome attachment site, maps near the origin of human mitochondrial DNA replication. J. Mol. Biol 150, 303–314 (1981). - PubMed
-
- Clayton DA Replication of animal mitochondrial DNA. Cell 28, 693–705 (1982). - PubMed
Methods-only references
-
- Jha A et al. Fibertools: fast and accurate DNA-m6A calling using single-molecule long-read sequencing. bioRxiv 2023.04.20.537673 (2023) doi:10.1101/2023.04.20.537673. - DOI
-
- TD buffer. Cold Spring Harb. Protoc. 2010, db.rec12138 (2010).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

