Baseline ctDNA gene alterations as a biomarker of survival after panitumumab and chemotherapy in metastatic colorectal cancer
- PMID: 38347302
- PMCID: PMC10957476
- DOI: 10.1038/s41591-023-02791-w
Baseline ctDNA gene alterations as a biomarker of survival after panitumumab and chemotherapy in metastatic colorectal cancer
Abstract
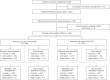
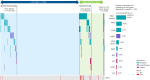
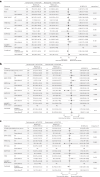
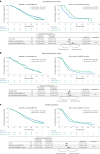
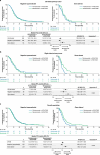
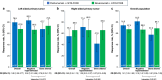
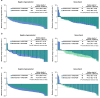
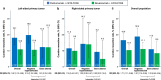
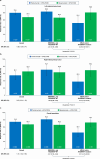
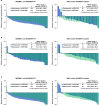
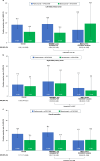
Certain genetic alterations and right-sided primary tumor location are associated with resistance to anti-epidermal growth factor (EGFR) treatment in metastatic colorectal cancer (mCRC). The phase 3 PARADIGM trial (n = 802) demonstrated longer overall survival with first-line anti-EGFR (panitumumab) versus antivascular endothelial growth factor (bevacizumab) plus modified FOLFOX6 in patients with RAS wild-type mCRC with left-sided primary tumors. This prespecified exploratory biomarker analysis of PARADIGM (n = 733) evaluated the association between circulating tumor DNA (ctDNA) gene alterations and efficacy outcomes, focusing on a broad panel of gene alterations associated with resistance to EGFR inhibition, including KRAS, NRAS, PTEN and extracellular domain EGFR mutations, HER2 and MET amplifications, and ALK, RET and NTRK1 fusions. Overall survival was prolonged with panitumumab plus modified FOLFOX6 versus bevacizumab plus modified FOLFOX6 in patients with ctDNA that lacked gene alterations in the panel (that is, negative hyperselected; median in the overall population: 40.7 versus 34.4 months; hazard ratio, 0.76; 95% confidence interval, 0.62-0.92) but was similar or inferior with panitumumab in patients with ctDNA that contained any gene alteration in the panel (19.2 versus 22.2 months; hazard ratio, 1.13; 95% confidence interval, 0.83-1.53), regardless of tumor sidedness. Negative hyperselection using ctDNA may guide optimal treatment selection in patients with mCRC. ClinicalTrials.gov registrations: NCT02394834 and NCT02394795 .
© 2024. The Author(s).
Conflict of interest statement
K.S.: Personal fees for consulting and advisory role from Bristol-Myers Squibb, Takeda, Ono Pharmaceutical, Novartis, Daiichi Sankyo, Amgen, Boehringer Ingelheim, Merck Pharmaceutical, Astellas, Guardant Health Japan, Janssen, AstraZeneca, Zymeworks Biopharmaceuticals, ALX Oncology and Bayer; honoraria from Bristol-Myers Squibb, Ono Pharmaceutical, Janssen, Eli Lilly, Astellas and AstraZeneca; research funding (all to institution) from Astellas, Ono Pharmaceutical, Daiichi Sankyo, Taiho Pharmaceutical, Chugai, Merck Pharmaceutical, Amgen, Eisai, PRA Health Sciences and Syneos Health, outside of the submitted work. K. Muro: Consulting and advisory role for Chugai Pharma, AstraZeneca, Ono Pharmaceutical and Amgen; honoraria from Chugai Pharma, Ono Pharmaceutical, Takeda, Eli Lilly, Bayer, Sanofi, Bristol-Myers Squibb and Taiho Pharmaceutical; research grant from Taiho Pharmaceutical, Astellas Pharma, Amgen Astellas Biopharma, (rest to institution) Merck Sharp & Dohme, Daiichi Sankyo, Shionogi, Kyowa Kirin, Gilead Sciences, Merck Serono, Pfizer, Sanofi, PAREXEL, Mediscience Planning, Sumitomo Dainippon Pharma, Solasia Pharma and Ono Pharmaceutical. J.W.: Speakers bureau for Covidien Japan, Johnson & Johnson/Janssen, Eli Lilly Japan and Takeda; research funding (to institution) from Medtronic, TERUMO and Stryker Japan. K. Yamazaki: Honoraria from Chugai Pharma, Daiichi Sankyo, Yakult Honsha, Takeda, Bayer, Merck Serono, Taiho Pharmaceutical, Eli Lilly, Sanofi, Ono Pharmaceutical, Merck Sharp & Dohme and Bristol-Myers Squibb; research funding (to institution) from Taiho Pharmaceutical. H. Ohori: Honoraria from Daiichi Sankyo, Yakult Honsha, Takeda, Taiho Pharmaceutical, Eli Lilly, Merck Sharp & Dohme and Bristol-Myers Squibb. N.A., M.Y., H. Ojima, Y.Y., K. Miwa, H.Y., K.A. and A.O. report having no relationships to disclose. A.T.: Grants from Takeda, Ono Pharmaceutical, Merck Sharp & Dohme, Bristol-Myers Squibb, Isofol Medical AB, Hutchison Medipharma, Incyte Corporation, Pfizer, Daiichi Sankyo; personal fees from Eli Lilly Japan, Taiho Pharmaceutical, Ono Pharmaceutical, Chugai Pharmaceutical, Takeda, Merck Serono and Merck Sharp & Dohme. A.M.: Personal fees from Eli Lilly Japan, Taiho Pharmaceutical, Ono Pharmaceutical, Bristol-Myers Squibb and Daiichi Sankyo. M.S.: Honoraria from Yakult Honsha, Takeda, Merck Serono, Taiho Pharmaceutical, Eli Lilly, Ono Pharmaceutical and Johnson & Johnson. E.O.: Speakers bureau for Chugai Pharma, Eli Lilly Japan, Takeda, Ono Pharmaceutical, Bayer Yakuhin and Bristol-Myers Squibb Japan. T.S.: Consulting and advisory role for Takeda; speakers bureau for Chugai Pharma, Eli Lilly Japan, Taiho Oncology, Takeda, Bayer Yakuhin, Ono Yakuhin and Daiichi Sankyo/UCB Japan. T.N.: Honoraria from Chugai Pharma, Taiho Pharmaceutical, Kaken Pharmaceutical, Daiichi Sankyo, Eli Lilly Japan, Takeda, Merck, Bayer and Boehringer Ingelheim; research funding (all to institution) from Chugai Pharma, Taiho Pharmaceutical, Kaken Pharmaceutical, Daiichi Sankyo and Eli Lilly Japan. Y.K.: Speakers bureau for Ono Pharmaceutical, Taiho, Chugai, Eli Lilly and Bayer Yakuhin; research funding from Ono Pharmaceutical, Taiho, Daiichi Sankyo, Chugai and IQVIA. T.K.: Honoraria from Chugai Pharma, Yakult Honsha, Ono Pharmaceutical, Takeda, Eli Lilly Japan, Taiho Pharmaceutical and Asahi Kasei; research funding from Chugai Pharma. I.M.: Employment with Takeda Pharmaceutical Company Ltd. K. Yamanaka: Employment with Takeda Pharmaceutical Company Ltd. M.H.: Employment with Takeda Pharmaceutical Company Ltd. J.S.: Employment with Takeda Pharmaceutical Company Ltd. T.M.: Honoraria from Chugai Pharma, AstraZeneca and Mivarisan. K. Yamamoto: Honoraria from Chugai Pharma, J-Pharma, Johokiko, Triceps and CMIC Holdings; research funding from Taiho, Boehringer Ingelheim, Takeda, Daiichi Sankyo and Astellas. R.Y.: Honoraria from Chugai Pharma, Takeda and BitBiome; consulting and advisory role for Takeda. H.U.: Speakers bureau for Takeda, Chugai Pharma and Taiho. K.T.: Honoraria from Chugai Pharma, Novartis, Takeda, Miyarisan Pharmaceutical, Bristol-Myers Squibb Japan, AstraZeneca, Illumina, Eisai, Boehringer Ingelheim Seiyaku and Bayer Yakuhin. T.Y.: Honoraria from Chugai Pharma, Merck, Bayer Yakuhin, Ono Pharmaceutical, Takeda, and Merck Sharp & Dohme; consulting and advisory role for Sumitomo Corp.; research funding (all to institution) from Merck Sharp & Dohme, Daiichi Sankyo, Ono Pharmaceutical, Taiho Pharmaceutical, Amgen, Sanofi, Pfizer, Genomedia, Sysmex, Nippon Boehringer Ingelheim, Eisai, FALCO Biosystems, Roche Diagnostics and Chugai Pharma.
Figures


References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

