Recombination between non-structural and structural genes as a mechanism of selection in lagoviruses: The evolutionary dead-end of an RHDV2 isolated from European hare
- PMID: 38347757
- PMCID: PMC10654597
- DOI: 10.1016/j.virusres.2023.199257
Recombination between non-structural and structural genes as a mechanism of selection in lagoviruses: The evolutionary dead-end of an RHDV2 isolated from European hare
Abstract
The genus Lagovirus, belonging to the family Caliciviridae, emerged around the 1980s. It includes highly pathogenic species, rabbit hemorrhagic disease virus (RHDV/GI.1) and European brown hare syndrome virus (EBHSV/GII.1), which cause fatal hepatitis, and nonpathogenic viruses with enteric tropism, rabbit calicivirus (RCV/GI.3,4) and hare calicivirus (HaCV/GII.2). Lagoviruses have evolved along two independent genetic lineages: GI (RHDV and RCV) in rabbits and GII (EBHSV and HaCV) in hares. To be emphasized is that genomes of lagoviruses, like other caliciviruses, are highly conserved at RdRp-VP60 junctions, favoring intergenotypic recombination events at this point. The recombination between an RCV (genotype GI.3), donor of non-structural (NS) genes, and an unknown virus, donor of structural (S) genes, likely led to the emergence of a new lagovirus in the European rabbit, called RHDV type 2 (GI.2), identified in Europe in 2010. New RHDV2 intergenotypic recombinants isolated in rabbits in Europe and Australia originated from similar events between RHDV2 (GI.2) and RHDV (GI.1) or RCV (GI.3,4). RHDV2 (GI.2) rapidly spread worldwide, replacing RHDV and showing several lagomorph species as secondary hosts. The recombination events in RHDV2 viruses have led to a number of viruses with very different combinations of NS and S genes. Recombinant RHDV2 with NS genes from hare lineage (GII) was recently identified in the European hare. This study investigated the first RHDV2 (GI.2) identified in Italy in European hare (RHDV2_Bg12), demonstrating that it was a new virus that originated from the recombination between RHDV2, as an S-gene donor and a hare lagovirus, not yet identified but presumably nonpathogenic, as an NS gene donor. When rabbits were inoculated with RHDV2_Bg12, neither deaths nor seroconversions were recorded, demonstrating that RHDV2_Bg12 cannot infect the rabbit. Furthermore, despite intensive and continuous field surveillance, RHDV2_Bg12 has never again been identified in either hares or rabbits in Italy or elsewhere. This result showed that the host specificity of lagoviruses can depend not only on S genes, as expected until today, but potentially also on some species-specific NS gene sequences. Therefore, because RHDV2 (GI.2) infects several lagomorphs, which in turn probably harbor several specific nonpathogenic lagoviruses, the possibility of new speciation, especially in those other than rabbits, is real. RHDV2 Bg_12 demonstrated this, although the attempt apparently failed.
Keywords: Animal experiments; Lagovirus; RHDV2; Recombination; Virus selection.
Copyright © 2023. Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




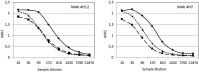
 , the liver of the Hare_Bcn14
, the liver of the Hare_Bcn14  , and the liver of the Hare_Bg12
, and the liver of the Hare_Bg12  . Abscissae indicate the inverse of the dilutions of the homogenates (40 means 1/40), whereas ordinates indicate the OD values (absorbance 492 nm = A492).
. Abscissae indicate the inverse of the dilutions of the homogenates (40 means 1/40), whereas ordinates indicate the OD values (absorbance 492 nm = A492).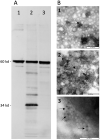
Similar articles
-
Genetic Characteristics and Phylogeographic Dynamics of Lagoviruses, 1988-2021.Viruses. 2023 Mar 23;15(4):815. doi: 10.3390/v15040815. Viruses. 2023. PMID: 37112796 Free PMC article.
-
Potent Protease Inhibitors of Highly Pathogenic Lagoviruses: Rabbit Hemorrhagic Disease Virus and European Brown Hare Syndrome Virus.Microbiol Spectr. 2022 Aug 31;10(4):e0014222. doi: 10.1128/spectrum.00142-22. Epub 2022 Jun 29. Microbiol Spectr. 2022. PMID: 35766511 Free PMC article.
-
Widespread occurrence of the non-pathogenic hare calicivirus (HaCV Lagovirus GII.2) in captive-reared and free-living wild hares in Europe.Transbound Emerg Dis. 2021 Mar;68(2):509-518. doi: 10.1111/tbed.13706. Epub 2020 Jul 14. Transbound Emerg Dis. 2021. PMID: 32603021 Free PMC article.
-
Characterisation of Lagovirus europaeus GI-RHDVs (Rabbit Haemorrhagic Disease Viruses) in Terms of Their Pathogenicity and Immunogenicity.Int J Mol Sci. 2024 May 14;25(10):5342. doi: 10.3390/ijms25105342. Int J Mol Sci. 2024. PMID: 38791380 Free PMC article. Review.
-
Rabbit hemorrhagic disease virus 2, 2010-2023: a review of global detections and affected species.J Vet Diagn Invest. 2024 Sep;36(5):617-637. doi: 10.1177/10406387241260281. Epub 2024 Jun 19. J Vet Diagn Invest. 2024. PMID: 39344909 Free PMC article. Review.
Cited by
-
Detection of a New Recombinant Rabbit Hemorrhagic Disease Virus 2 in China and Development of Virus-like Particle-Based Vaccine.Viruses. 2025 May 16;17(5):710. doi: 10.3390/v17050710. Viruses. 2025. PMID: 40431721 Free PMC article.
-
Two decades of occurrence of non-pathogenic rabbit lagoviruses in Italy and their genomic characterization.Sci Rep. 2024 Nov 25;14(1):29234. doi: 10.1038/s41598-024-79670-y. Sci Rep. 2024. PMID: 39587141 Free PMC article.
-
Comment on Shah et al. Genetic Characteristics and Phylogeographic Dynamics of Lagoviruses, 1988-2021. Viruses 2023, 15, 815.Viruses. 2024 Jun 7;16(6):927. doi: 10.3390/v16060927. Viruses. 2024. PMID: 38932219 Free PMC article.
-
Identification and Characterisation of a Myxoma Virus Detected in the Italian Hare (Lepus corsicanus).Viruses. 2024 Mar 12;16(3):437. doi: 10.3390/v16030437. Viruses. 2024. PMID: 38543802 Free PMC article.
-
Domestic European Rabbits Oryctolagus cuniculus: A Super-Highway for the Spread of Emergent Viral Diseases to Other Lagomorphs?Transbound Emerg Dis. 2025 Jun 6;2025:1129135. doi: 10.1155/tbed/1129135. eCollection 2025. Transbound Emerg Dis. 2025. PMID: 40521303 Free PMC article.
References
-
- Abrantes J., Lopes A.M., Lemaitre E., Ahola H., Banihashem F., Droillard C., et al. Retrospective analysis shows that most RHDV GI.1 strains circulating since the late 1990s in France and Sweden were recombinant GI.3P-GI.1d strains. Genes (Basel) 2020;11:910. doi: 10.3390/genes11080910. - DOI - PMC - PubMed
-
- Asin J., Nyaoke A.C., Moore J.D., Gonzalez-Astudillo V., Clifford D.L., Lantz E.L., et al. Outbreak of rabbit hemorrhagic disease virus 2 in the southwestern United States: first detections in southern California. J. Vet. Diagn. Invest. 2021;33:728–731. doi: 10.1177/10406387211006353. - DOI - PMC - PubMed
-
- Asin J., Rejmanek D., Clifford D.L., Mikolon A.B., Henderson E.E., Nyaoke A.C., et al. Early circulation of rabbit haemorrhagic disease virus type 2 in domestic and wild lagomorphs in southern California, USA (2020-2021) Transbound Emerg. Dis. 2022;69:e394–e405. doi: 10.1111/tbed.14315. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

