Identification and characterization of CLEC11A and its derived immune signature in gastric cancer
- PMID: 38348052
- PMCID: PMC10859539
- DOI: 10.3389/fimmu.2024.1324959
Identification and characterization of CLEC11A and its derived immune signature in gastric cancer
Abstract
Introduction: C-type lectin domain family 11 member A (CLEC11A) was characterized as a growth factor that mainly regulates hematopoietic function and differentiation of bone cells. However, the involvement of CLEC11A in gastric cancer (GC) is not well understood.
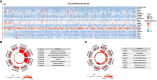
Methods: Transcriptomic data and clinical information pertaining to GC were obtained and analyzed from publicly available databases. The relationships between CLEC11A and prognoses, genetic alterations, tumor microenvironment (TME), and therapeutic responses in GC patients were analyzed by bioinformatics methods. A CLEC11A-derived immune signature was developed and validated, and its mutational landscapes, immunological characteristics as well as drug sensitivities were explored. A nomogram was established by combining CLEC11A-derived immune signature and clinical factors. The expression and carcinogenic effects of CLEC11A in GC were verified by qRT-PCR, cell migration, invasion, cell cycle analysis, and in vivo model analysis. Myeloid-derived suppressor cells (MDSCs), regulatory T cells (Tregs), M2 macrophages, and T cells in tumor samples extracted from mice were analyzed utilizing flow cytometry analysis.
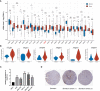
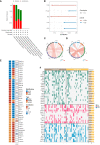
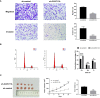
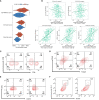
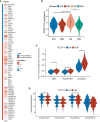
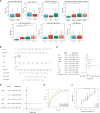
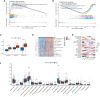
Results: CLEC11A was over-expressed in GC, and the elevated CLEC11A expression indicated an unfavorable prognosis in GC patients. CLEC11A was involved in genomic alterations and associated with the TME in GC. Moreover, elevated CLEC11A was found to reduce the benefit of immunotherapy according to immunophenoscore (IPS) and the tumor immune dysfunction, exclusion (TIDE). After validation, the CLEC11A-derived immune signature demonstrated a consistent ability to predict the survival outcomes in GC patients. A nomogram that quantifies survival probability was constructed to improve the accuracy of prognosis prediction in GC patients. Using shRNA to suppress the expression of CLEC11A led to significant inhibitions of cell cycle progression, migration, and invasion, as well as a marked reduction of in vivo tumor growth. Moreover, the flow cytometry assay showed that the knock-down of CLEC11A increased the infiltration of cytotoxic CD8+ T cells and helper CD4+ T into tumors while decreasing the percentage of M2 macrophages, MDSCs, and Tregs.
Conclusion: Collectively, our findings revealed that CLEC11A could be a prognostic and immunological biomarker in GC, and CLEC11A-derived immune signature might serve as a new option for clinicians to predict outcomes and formulate personalized treatment plans for GC patients.
Keywords: Clec11a; gastric cancer; immunotherapy; prognosis; tumor microenvironment.
Copyright © 2024 Zheng, Gong, Li, Cheng, Luo, Huang and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
CLEC11A expression as a prognostic biomarker in correlation to immune cells of gastric cancer.Biomol Biomed. 2024 Jan 3;24(1):101-124. doi: 10.17305/bb.2023.9384. Biomol Biomed. 2024. PMID: 37597212 Free PMC article.
-
Identification and validation of a dysregulated TME-related gene signature for predicting prognosis, and immunological properties in bladder cancer.Front Immunol. 2023 Oct 27;14:1213947. doi: 10.3389/fimmu.2023.1213947. eCollection 2023. Front Immunol. 2023. PMID: 37965307 Free PMC article.
-
A nomogram model based on the number of examined lymph nodes-related signature to predict prognosis and guide clinical therapy in gastric cancer.Front Immunol. 2022 Nov 2;13:947802. doi: 10.3389/fimmu.2022.947802. eCollection 2022. Front Immunol. 2022. PMID: 36405735 Free PMC article.
-
A novel necroptosis-related gene index for predicting prognosis and a cold tumor immune microenvironment in stomach adenocarcinoma.Front Immunol. 2022 Oct 27;13:968165. doi: 10.3389/fimmu.2022.968165. eCollection 2022. Front Immunol. 2022. PMID: 36389725 Free PMC article. Review.
-
Regulation of regulatory T cells and tumor-associated macrophages in gastric cancer tumor microenvironment.Cancer Med. 2024 Jan;13(2):e6959. doi: 10.1002/cam4.6959. Cancer Med. 2024. PMID: 38349050 Free PMC article. Review.
Cited by
-
A New Medical Evaluation for Gastric Cancer Patients to Increase the Success Rate of Immunotherapy: A 2024 Update.Pharmaceuticals (Basel). 2024 Aug 24;17(9):1121. doi: 10.3390/ph17091121. Pharmaceuticals (Basel). 2024. PMID: 39338286 Free PMC article. Review.
-
Predictive Factors of Immunotherapy in Gastric Cancer: A 2024 Update.Diagnostics (Basel). 2024 Jun 13;14(12):1247. doi: 10.3390/diagnostics14121247. Diagnostics (Basel). 2024. PMID: 38928662 Free PMC article. Review.
-
Exosomes derived from minor salivary gland mesenchymal stem cells: a promising novel exosome exhibiting pro-angiogenic and wound healing effects similar to those of adipose-derived stem cell exosomes.Stem Cell Res Ther. 2024 Dec 3;15(1):462. doi: 10.1186/s13287-024-04069-5. Stem Cell Res Ther. 2024. PMID: 39627883 Free PMC article.
-
Revealing the mechanisms of RAC3 in tumor aggressiveness, the immunotherapy response, and drug resistance in bladder cancer.Front Oncol. 2024 Sep 16;14:1466319. doi: 10.3389/fonc.2024.1466319. eCollection 2024. Front Oncol. 2024. PMID: 39351351 Free PMC article.
-
Potential predictive value of CD8A and PGF protein expression in gastric cancer patients treated with neoadjuvant immunotherapy.BMC Cancer. 2025 Apr 12;25(1):674. doi: 10.1186/s12885-025-14046-7. BMC Cancer. 2025. PMID: 40221689 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

