Evaluation of 19 years of international external proficiency testing for high-resolution HLA typing
- PMID: 38348410
- PMCID: PMC10859402
- DOI: 10.3389/fgene.2023.1290915
Evaluation of 19 years of international external proficiency testing for high-resolution HLA typing
Abstract
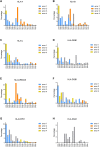
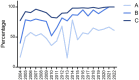
The international high-resolution external proficiency testing (EPT) started in 2004 with high-resolution typing of human leucocyte antigen (HLA) class I (HLA-A,B,C) and HLA class II (HLA-DRB1, DRB345, DQB1, and DPB1) alleles, since possibilities for such an EPT within Europe were limited and all existing EPTs at that time made use of the comparison of HLA typing results without a reference. This EPT was set up as a collaboration between the HLA laboratory of Leiden, providing DNA samples to the participants, and the laboratory of Maastricht, performing the high-resolution typing as the reference result and evaluating the results of all participants according to the prevailing European Federation for Immunogenetics (EFI) standards. Once a year, 12 samples were sent to the participating laboratories, and evaluation and certificates were provided at the end of that same year. During the years, the EPT was extended to low-resolution HLA class I and II typing, high-resolution typing including DQA1 and DPA1, and allelic resolution typing for HLA class I, the latter one being unique in this field. Evaluation of the high-resolution typing results of the last 19 years showed a clear increase in the number of loci tested by the participating laboratories and a clear change of method from Sanger sequencing with additional other techniques (SSO/SSP) to the nowadays widely used next-generation sequencing method. By strictly using the EFI rules for high-resolution HLA typing, the participants were made aware of the ambiguities within exons 2 and 3 for class I and exon 2 for class II and the presence of null alleles even in a two-field HLA typing. There was an impressive learning curve, resulting in >98% correctly typed samples since 2017 and a 100% fulfillment of EFI rules for all laboratories for all loci submitted in the last 2 years. Overall, this EPT meets the need of an EPT for high-resolution typing for EFI accreditation.
Keywords: HLA high-resolution typing; NGS; accreditation; histocompatibility; immunogenetic tests; quality assessment; quality control; sequencing.
Copyright © 2024 Voorter, Groeneveld, Heidt and Wieten.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Development of the HLA proficiency testing for Central and East Europe.Int J Immunogenet. 2008 Aug;35(4-5):409-16. doi: 10.1111/j.1744-313X.2008.00789.x. Int J Immunogenet. 2008. PMID: 18976448
-
Human leukocyte antigen proficiency testing for Central and Eastern Europe: a summary of over 10 years of activity.Transplant Proc. 2010 Oct;42(8):3263-5. doi: 10.1016/j.transproceed.2010.07.020. Transplant Proc. 2010. PMID: 20970668 Review.
-
Extended genomic HLA typing identifies previously unrecognized mismatches in living kidney transplantation.Front Immunol. 2023 Jan 27;14:1094862. doi: 10.3389/fimmu.2023.1094862. eCollection 2023. Front Immunol. 2023. PMID: 36776892 Free PMC article.
-
A national proficiency scheme for human leucocyte antigen typing by next-generation sequencing.Clin Chim Acta. 2022 Aug 1;533:85-88. doi: 10.1016/j.cca.2022.06.015. Epub 2022 Jun 20. Clin Chim Acta. 2022. PMID: 35738456
-
Quality control project of NGS HLA genotyping for the 17th International HLA and Immunogenetics Workshop.Hum Immunol. 2019 Apr;80(4):228-236. doi: 10.1016/j.humimm.2019.01.009. Epub 2019 Feb 6. Hum Immunol. 2019. PMID: 30738112 Free PMC article. Review.
Cited by
-
Ten years of the immunogenetics laboratory performance assessment programme and its impact on the donor and transplant network.Biomedica. 2024 Dec 23;44(Sp. 2):155-167. doi: 10.7705/biomedica.7589. Biomedica. 2024. PMID: 39836833 Free PMC article. English, Spanish.
References
-
- Voorter C. E., Groeneweg M., Groeneveld L., Tilanus M. G. (2016). Uncommon HLA alleles identified by hemizygous ultra-high Sanger sequencing: haplotype associations and reconsideration of their assignment in the Common and Well-Documented catalogue. Hum. Immunol. 77 (2), 184–190. 10.1016/j.humimm.2015.11.016 - DOI - PubMed
-
- Voorter C. E. M., Palusci F., Tilanus M. G. J. (2014). “Sequence-based typing of HLA: an improved group-specific full-length gene sequencing approach,” in Methods mol biol. Editor Beksac M. (Totowa, NJ: Humana Press; ), 101–114. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

