Standardizing Extracted Data Using Automated Application of Controlled Vocabularies
- PMID: 38349723
- PMCID: PMC10863721
- DOI: 10.1289/EHP13215
Standardizing Extracted Data Using Automated Application of Controlled Vocabularies
Abstract
Background: Extraction of toxicological end points from primary sources is a central component of systematic reviews and human health risk assessments. To ensure optimal use of these data, consistent language should be used for end point descriptions. However, primary source language describing treatment-related end points can vary greatly, resulting in large labor efforts to manually standardize extractions before data are fit for use.
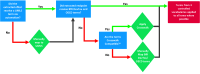
Objectives: To minimize these labor efforts, we applied an augmented intelligence approach and developed automated tools to support standardization of extracted information via application of preexisting controlled vocabularies.
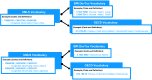
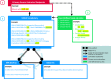
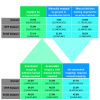
Methods: We created and applied a harmonized controlled vocabulary crosswalk, consisting of Unified Medical Language System (UMLS) codes, German Federal Institute for Risk Assessment (BfR) DevTox harmonized terms, and The Organization for Economic Co-operation and Development (OECD) end point vocabularies, to roughly 34,000 extractions from prenatal developmental toxicology studies conducted by the National Toxicology Program (NTP) and 6,400 extractions from European Chemicals Agency (ECHA) prenatal developmental toxicology studies, all recorded based on the original study report language.
Results: We automatically applied standardized controlled vocabulary terms to 75% of the NTP extracted end points and 57% of the ECHA extracted end points. Of all the standardized extracted end points, about half (51%) required manual review for potential extraneous matches or inaccuracies. Extracted end points that were not mapped to standardized terms tended to be too general or required human logic to find a good match. We estimate that this augmented intelligence approach saved hours of manual effort and yielded valuable resources including a controlled vocabulary crosswalk, organized related terms lists, code for implementing an automated mapping workflow, and a computationally accessible dataset.
Discussion: Augmenting manual efforts with automation tools increased the efficiency of producing a findable, accessible, interoperable, and reusable (FAIR) dataset of regulatory guideline studies. This open-source approach can be readily applied to other legacy developmental toxicology datasets, and the code design is customizable for other study types. https://doi.org/10.1289/EHP13215.
Figures


Similar articles
-
Evaluating the coverage of controlled health data terminologies: report on the results of the NLM/AHCPR large scale vocabulary test.J Am Med Inform Assoc. 1997 Nov-Dec;4(6):484-500. doi: 10.1136/jamia.1997.0040484. J Am Med Inform Assoc. 1997. PMID: 9391936 Free PMC article.
-
Features of a FAIR vocabulary.J Biomed Semantics. 2023 Jun 1;14(1):6. doi: 10.1186/s13326-023-00286-8. J Biomed Semantics. 2023. PMID: 37264430 Free PMC article.
-
The future of Cochrane Neonatal.Early Hum Dev. 2020 Nov;150:105191. doi: 10.1016/j.earlhumdev.2020.105191. Epub 2020 Sep 12. Early Hum Dev. 2020. PMID: 33036834
-
Behavioural modification interventions for medically unexplained symptoms in primary care: systematic reviews and economic evaluation.Health Technol Assess. 2020 Sep;24(46):1-490. doi: 10.3310/hta24460. Health Technol Assess. 2020. PMID: 32975190 Free PMC article.
-
Modelling approaches for histology-independent cancer drugs to inform NICE appraisals: a systematic review and decision-framework.Health Technol Assess. 2021 Dec;25(76):1-228. doi: 10.3310/hta25760. Health Technol Assess. 2021. PMID: 34990339
Cited by
-
Artificial intelligence (AI)-it's the end of the tox as we know it (and I feel fine).Arch Toxicol. 2024 Mar;98(3):735-754. doi: 10.1007/s00204-023-03666-2. Epub 2024 Jan 20. Arch Toxicol. 2024. PMID: 38244040 Free PMC article. Review.
References
-
- WHO (World Health Organization). 2021. Framework for the Use of Systematic Review in Chemical Risk Assessment. Geneva, Switzerland: WHO, Chemical Safety and Health Unit. https://www.who.int/publications/i/item/9789240034488 [accessed 8 October 2021].
-
- Hood RD. 2016. Developmental and Reproductive Toxicology: A Practical Approach. Boca Raton, FL: CRC Press.
-
- US EPA (US Environmental Protection Agency). 1991. Guidelines for developmental toxicity risk assessment. Fed Reg 56(234):63798–63826.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

