Learning to Make Rare and Complex Diagnoses With Generative AI Assistance: Qualitative Study of Popular Large Language Models
- PMID: 38349725
- PMCID: PMC10900078
- DOI: 10.2196/51391
Learning to Make Rare and Complex Diagnoses With Generative AI Assistance: Qualitative Study of Popular Large Language Models
Abstract
Background: Patients with rare and complex diseases often experience delayed diagnoses and misdiagnoses because comprehensive knowledge about these diseases is limited to only a few medical experts. In this context, large language models (LLMs) have emerged as powerful knowledge aggregation tools with applications in clinical decision support and education domains.
Objective: This study aims to explore the potential of 3 popular LLMs, namely Bard (Google LLC), ChatGPT-3.5 (OpenAI), and GPT-4 (OpenAI), in medical education to enhance the diagnosis of rare and complex diseases while investigating the impact of prompt engineering on their performance.
Methods: We conducted experiments on publicly available complex and rare cases to achieve these objectives. We implemented various prompt strategies to evaluate the performance of these models using both open-ended and multiple-choice prompts. In addition, we used a majority voting strategy to leverage diverse reasoning paths within language models, aiming to enhance their reliability. Furthermore, we compared their performance with the performance of human respondents and MedAlpaca, a generative LLM specifically designed for medical tasks.
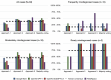
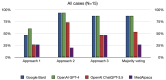
Results: Notably, all LLMs outperformed the average human consensus and MedAlpaca, with a minimum margin of 5% and 13%, respectively, across all 30 cases from the diagnostic case challenge collection. On the frequently misdiagnosed cases category, Bard tied with MedAlpaca but surpassed the human average consensus by 14%, whereas GPT-4 and ChatGPT-3.5 outperformed MedAlpaca and the human respondents on the moderately often misdiagnosed cases category with minimum accuracy scores of 28% and 11%, respectively. The majority voting strategy, particularly with GPT-4, demonstrated the highest overall score across all cases from the diagnostic complex case collection, surpassing that of other LLMs. On the Medical Information Mart for Intensive Care-III data sets, Bard and GPT-4 achieved the highest diagnostic accuracy scores, with multiple-choice prompts scoring 93%, whereas ChatGPT-3.5 and MedAlpaca scored 73% and 47%, respectively. Furthermore, our results demonstrate that there is no one-size-fits-all prompting approach for improving the performance of LLMs and that a single strategy does not universally apply to all LLMs.
Conclusions: Our findings shed light on the diagnostic capabilities of LLMs and the challenges associated with identifying an optimal prompting strategy that aligns with each language model's characteristics and specific task requirements. The significance of prompt engineering is highlighted, providing valuable insights for researchers and practitioners who use these language models for medical training. Furthermore, this study represents a crucial step toward understanding how LLMs can enhance diagnostic reasoning in rare and complex medical cases, paving the way for developing effective educational tools and accurate diagnostic aids to improve patient care and outcomes.
Keywords: AI assistance; Bard; ChatGPT 3.5; GPT-4; MedAlpaca; artificial intelligence; clinical decision support; complex diagnosis; complex diseases; consistency; language model; medical education; medical training; natural language processing; prediction model; prompt engineering; rare diseases; reliability.
©Tassallah Abdullahi, Ritambhara Singh, Carsten Eickhoff. Originally published in JMIR Medical Education (https://mededu.jmir.org), 13.02.2024.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures
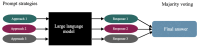
Similar articles
-
Evaluating Large Language Models for the National Premedical Exam in India: Comparative Analysis of GPT-3.5, GPT-4, and Bard.JMIR Med Educ. 2024 Feb 21;10:e51523. doi: 10.2196/51523. JMIR Med Educ. 2024. PMID: 38381486 Free PMC article.
-
An Empirical Evaluation of Prompting Strategies for Large Language Models in Zero-Shot Clinical Natural Language Processing: Algorithm Development and Validation Study.JMIR Med Inform. 2024 Apr 8;12:e55318. doi: 10.2196/55318. JMIR Med Inform. 2024. PMID: 38587879 Free PMC article.
-
Performance of Large Language Models (ChatGPT, Bing Search, and Google Bard) in Solving Case Vignettes in Physiology.Cureus. 2023 Aug 4;15(8):e42972. doi: 10.7759/cureus.42972. eCollection 2023 Aug. Cureus. 2023. PMID: 37671207 Free PMC article.
-
Artificial Intelligence for Anesthesiology Board-Style Examination Questions: Role of Large Language Models.J Cardiothorac Vasc Anesth. 2024 May;38(5):1251-1259. doi: 10.1053/j.jvca.2024.01.032. Epub 2024 Feb 1. J Cardiothorac Vasc Anesth. 2024. PMID: 38423884 Review.
-
The Role of Large Language Models in Transforming Emergency Medicine: Scoping Review.JMIR Med Inform. 2024 May 10;12:e53787. doi: 10.2196/53787. JMIR Med Inform. 2024. PMID: 38728687 Free PMC article.
Cited by
-
Patient-Representing Population's Perceptions of GPT-Generated Versus Standard Emergency Department Discharge Instructions: Randomized Blind Survey Assessment.J Med Internet Res. 2024 Aug 2;26:e60336. doi: 10.2196/60336. J Med Internet Res. 2024. PMID: 39094112 Free PMC article. Clinical Trial.
-
Large language models in critical care.J Intensive Med. 2024 Dec 24;5(2):113-118. doi: 10.1016/j.jointm.2024.12.001. eCollection 2025 Apr. J Intensive Med. 2024. PMID: 40241839 Free PMC article. Review.
-
The large language model diagnoses tuberculous pleural effusion in pleural effusion patients through clinical feature landscapes.Respir Res. 2025 Feb 12;26(1):52. doi: 10.1186/s12931-025-03130-y. Respir Res. 2025. PMID: 39939874 Free PMC article.
-
Evaluating multimodal AI in medical diagnostics.NPJ Digit Med. 2024 Aug 7;7(1):205. doi: 10.1038/s41746-024-01208-3. NPJ Digit Med. 2024. PMID: 39112822 Free PMC article.
-
Toward Clinical Generative AI: Conceptual Framework.JMIR AI. 2024 Jun 7;3:e55957. doi: 10.2196/55957. JMIR AI. 2024. PMID: 38875592 Free PMC article.
References
-
- Introducing ChatGPT. OpenAI. [2023-03-23]. https://openai.com/blog/chatgpt/
-
- Manyika J, Hsiao S. An overview of Bard: an early experiment with generative AI. Google. [2024-01-26]. https://ai.google/static/documents/google-about-bard.pdf .
-
- Achiam J, Adler S, Agarwal S, Ahmad L, Akkaya I, Aleman FL, Almeida D, Altenschmidt J, Altman S, Anadkat S, Avila R, Babuschkin I, Balaji S, Balcom V, Baltescu P, Bao H, Bavarian M, Belgum J, Bello I, Berdine J, Bernadett-Shapiro G, Berner C, Bogdonoff L, Boiko O, Boyd M, Brakman AL, Brockman G, Brooks T, Brundage M, Button K, Cai T, Campbell R, Cann A, Carey B, Carlson C, Carmichael R, Chan B, Chang C, Chantzis F, Chen D, Chen S, Chen R, Chen J, Chen M, Chess B, Cho C, Chu C, Chung HW, Cummings D, Currier J, Dai Y, Decareaux C, Degry T, Deutsch N, Deville D, Dhar A, Dohan D, Dowling S, Dunning S, Ecoffet A, Eleti A, Eloundou T, Farhi D, Fedus L, Felix N, Fishman SP, Forte J, Fulford I, Gao L, Georges E, Gibson C, Goel V, Gogineni T, Goh G, Gontijo-Lopes R, Gordon J, Grafstein M, Gray S, Greene R, Gross J, Gu SS, Guo Y, Hallacy C, Han J, Harris J, He Y, Heaton M, Heidecke J, Hesse C, Hickey A, Hickey W, Hoeschele P, Houghton B, Hsu K, Hu S, Hu X, Huizinga J, Jain S, Jain S. GPT-4 technical report. arXiv. Preprint posted online March 15, 2023. https://arxiv.org/abs/2303.08774
-
- Kung TH, Cheatham M, Medenilla A, Sillos C, De Leon L, Elepaño C, Madriaga M, Aggabao R, Diaz-Candido G, Maningo J, Tseng V. Performance of ChatGPT on USMLE: potential for AI-assisted medical education using large language models. PLOS Digit Health. 2023 Feb 9;2(2):e0000198. doi: 10.1371/journal.pdig.0000198. https://europepmc.org/abstract/MED/36812645 PDIG-D-22-00371 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

