Long interspersed nuclear elements safeguard neural progenitors from precocious differentiation
- PMID: 38349791
- PMCID: PMC10948021
- DOI: 10.1016/j.celrep.2024.113774
Long interspersed nuclear elements safeguard neural progenitors from precocious differentiation
Abstract
Long interspersed nuclear element-1 (L1 or LINE-1) is a highly abundant mobile genetic element in both humans and mice, comprising almost 20% of each genome. L1s are silenced by several mechanisms, as their uncontrolled expression has the potential to induce genomic instability. However, L1s are paradoxically expressed at high levels in differentiating neural progenitor cells. Using in vitro and in vivo techniques to modulate L1 expression, we report that L1s play a critical role in both human and mouse brain development by regulating the rate of neural differentiation in a reverse-transcription-independent manner.
Keywords: CP: Developmental biology; CP: Neuroscience; L1; LINE-1; brain development; neural progenitor cells; repetitive elements.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
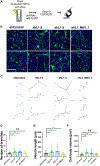

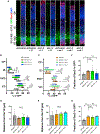
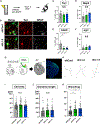
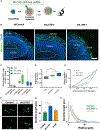
References
-
- Jönsson ME, Ludvik Brattås P, Gustafsson C, Petri R, Yudovich D, Pircs K, Verschuere S, Madsen S, Hansson J, Larsson J, et al. (2019). Activation of neuronal genes via LINE-1 elements upon global DNA demethylation in human neural progenitors. Nat. Commun 10, 3182. 10.1038/s41467-019-11150-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

