Genomic and transcriptomic profiling of peripheral T cell lymphoma reveals distinct molecular and microenvironment subtypes
- PMID: 38350451
- PMCID: PMC10897627
- DOI: 10.1016/j.xcrm.2024.101416
Genomic and transcriptomic profiling of peripheral T cell lymphoma reveals distinct molecular and microenvironment subtypes
Abstract
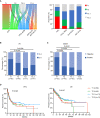
Peripheral T cell lymphoma (PTCL) is a heterogeneous group of non-Hodgkin's lymphomas varying in clinical, phenotypic, and genetic features. The molecular pathogenesis and the role of the tumor microenvironment in PTCL are poorly understood, with limited biomarkers available for genetic subtyping and targeted therapies. Through an integrated genomic and transcriptomic study of 221 PTCL patients, we delineate the genetic landscape of PTCL, enabling molecular and microenvironment classification. According to the mutational status of RHOA, TET2, histone-modifying, and immune-related genes, PTCL is divided into 4 molecular subtypes with discrete patterns of gene expression, biological aberrations, and vulnerabilities to targeted agents. We also perform an unsupervised clustering on the microenvironment transcriptional signatures and categorize PTCL into 4 lymphoma microenvironment subtypes based on characteristic activation of oncogenic pathways and composition of immune communities. Our findings highlight the potential clinical rationale of future precision medicine strategies that target both molecular and microenvironment alterations in PTCL.
Copyright © 2024. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






References
-
- Zain J.M., Hanona P. Aggressive T-cell lymphomas: 2021 updates on diagnosis, risk stratification and management. Am. J. Hematol. 2021;96:1027–1046. - PubMed
-
- Yoon S.E., Song Y., Kim S.J., Yoon D.H., Chen T.Y., Koh Y., Kang K.W., Lee H.S., Tay K.K.W., Lim S.T., et al. Comprehensive analysis of peripheral T-cell and natural killer/T-cell lymphoma in Asian patients: A multinational, multicenter, prospective registry study in Asia. Lancet Reg. Health. West. Pac. 2021;10 - PMC - PubMed
-
- Fiore D., Cappelli L.V., Broccoli A., Zinzani P.L., Chan W.C., Inghirami G. Peripheral T cell lymphomas: from the bench to the clinic. Nat. Rev. Cancer. 2020;20:323–342. - PubMed
-
- Teras L.R., DeSantis C.E., Cerhan J.R., Morton L.M., Jemal A., Flowers C.R. US lymphoid malignancy statistics by World Health Organization subtypes. CA A Cancer J. Clin. 2016;66:443–459. - PubMed
-
- Watatani Y., Sato Y., Miyoshi H., Sakamoto K., Nishida K., Gion Y., Nagata Y., Shiraishi Y., Chiba K., Tanaka H., et al. Molecular heterogeneity in peripheral T-cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia. 2019;33:2867–2883. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

