Regulatory sites of CaM-sensitive adenylyl cyclase AC8 revealed by cryo-EM and structural proteomics
- PMID: 38351373
- PMCID: PMC10933263
- DOI: 10.1038/s44319-024-00076-y
Regulatory sites of CaM-sensitive adenylyl cyclase AC8 revealed by cryo-EM and structural proteomics
Abstract
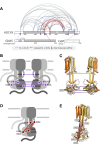
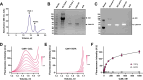
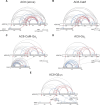
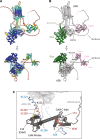
Membrane adenylyl cyclase AC8 is regulated by G proteins and calmodulin (CaM), mediating the crosstalk between the cAMP pathway and Ca2+ signalling. Despite the importance of AC8 in physiology, the structural basis of its regulation by G proteins and CaM is not well defined. Here, we report the 3.5 Å resolution cryo-EM structure of the bovine AC8 bound to the stimulatory Gαs protein in the presence of Ca2+/CaM. The structure reveals the architecture of the ordered AC8 domains bound to Gαs and the small molecule activator forskolin. The extracellular surface of AC8 features a negatively charged pocket, a potential site for unknown interactors. Despite the well-resolved forskolin density, the captured state of AC8 does not favour tight nucleotide binding. The structural proteomics approaches, limited proteolysis and crosslinking mass spectrometry (LiP-MS and XL-MS), allowed us to identify the contact sites between AC8 and its regulators, CaM, Gαs, and Gβγ, as well as to infer the conformational changes induced by these interactions. Our results provide a framework for understanding the role of flexible regions in the mechanism of AC regulation.
Keywords: Adenylyl cyclase; Calmodulin; Heterotrimeric G protein; Structural Proteomics; cryo-Electron Microscopy (cryo-EM).
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








Similar articles
-
Structural insights into calmodulin/adenylyl cyclase 8 interaction.Anal Bioanal Chem. 2013 Nov;405(29):9333-42. doi: 10.1007/s00216-013-7358-3. Epub 2013 Sep 27. Anal Bioanal Chem. 2013. PMID: 24071896
-
Identification of FDA-Approved Small Molecules Capable of Disrupting the Calmodulin-Adenylyl Cyclase 8 Interaction through Direct Binding to Calmodulin.ACS Chem Neurosci. 2018 Feb 21;9(2):346-357. doi: 10.1021/acschemneuro.7b00349. Epub 2017 Oct 16. ACS Chem Neurosci. 2018. PMID: 28968502 Free PMC article.
-
Separate elements within a single IQ-like motif in adenylyl cyclase type 8 impart ca2+/calmodulin binding and autoinhibition.J Biol Chem. 2009 Jun 5;284(23):15573-88. doi: 10.1074/jbc.M809585200. Epub 2009 Mar 21. J Biol Chem. 2009. PMID: 19305019 Free PMC article.
-
The adenylyl cyclase family.Mol Cell Biochem. 1991 May 29-Jun 12;104(1-2):73-9. doi: 10.1007/BF00229806. Mol Cell Biochem. 1991. PMID: 1656197 Review.
-
The structure, catalytic mechanism and regulation of adenylyl cyclase.Curr Opin Struct Biol. 1998 Dec;8(6):713-9. doi: 10.1016/s0959-440x(98)80090-0. Curr Opin Struct Biol. 1998. PMID: 9914249 Review.
Cited by
-
KCTD1 regulation of Adenylyl cyclase type 5 adjusts striatal cAMP signaling.Proc Natl Acad Sci U S A. 2024 Oct 22;121(43):e2406686121. doi: 10.1073/pnas.2406686121. Epub 2024 Oct 16. Proc Natl Acad Sci U S A. 2024. PMID: 39413138 Free PMC article.
-
Frontiers in integrative structural modeling of macromolecular assemblies.QRB Discov. 2025 Jan 22;6:e3. doi: 10.1017/qrd.2024.15. eCollection 2025. QRB Discov. 2025. PMID: 39944881 Free PMC article. Review.
-
Analysis of Limited Proteolysis-Coupled Mass Spectrometry Data.Mol Cell Proteomics. 2025 Apr;24(4):100934. doi: 10.1016/j.mcpro.2025.100934. Epub 2025 Mar 7. Mol Cell Proteomics. 2025. PMID: 40058498 Free PMC article.
References
-
- Barrett T, Dowle M, Srinivasan A, Gorecki J, Chirico M, Hocking T (2024) data.table: Extension of ‘data.frame’. R package version 1.14.99. https://Rdatatable.gitlab.io/data.table, https://github.com/Rdatatable/data.table, https://r-datatable.com
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

