Meta-analysis of microarray data to determine gene indicators involved in cisplatin resistance in non-small cell lung cancer
- PMID: 38351531
- PMCID: PMC10864718
- DOI: 10.1002/cnr2.1970
Meta-analysis of microarray data to determine gene indicators involved in cisplatin resistance in non-small cell lung cancer
Abstract
Background: Lung cancer is a major cause of cancer-related mortality worldwide, with a 5-year survival rate of approximately 22%. Cisplatin is one of the standard first-line chemotherapeutic agents for non-small cell lung cancer (NSCLC), but its efficacy is often limited by the development of resistance. Despite extensive research on the molecular mechanisms of chemoresistance, the underlying causes remain elusive and complex.
Aims: We analyzed three microarray datasets to find the gene signature and key pathways related to cisplatin resistance in NSCLC.
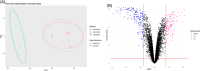
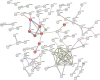
Methods and results: We compared the gene expression of sensitive and resistant NSCLC cell lines treated with cisplatin. We found 274 DEGs, including 111 upregulated and 163 downregulated genes, in the resistant group. Gene set enrichment analysis showed the potential roles of several DEGs, such as TUBB2B, MAPK7, TUBAL3, MAP2K5, SMUG1, NTHL1, PARP3, NTRK1, G6PD, PDK1, HEY1, YTHDF2, CD274, and MAGEA1, in cisplatin resistance. Functional analysis revealed the involvement of pathways, such as gap junction, base excision repair, central carbon metabolism, and Notch signaling in the resistant cell lines.
Conclusion: We identified several molecular factors that contribute to cisplatin resistance in NSCLC cell lines, involving genes and pathways that regulate gap junction communication, DNA damage repair, ROS balance, EMT induction, and stemness maintenance. These genes and pathways could be targets for future studies to overcome cisplatin resistance in NSCLC.
Keywords: BER; EMT; PARP3; cisplatin resistance; microarray; non-small cell lung cancer.
© 2024 The Authors. Cancer Reports published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Herbst RS, Morgensztern D, Boshoff C. The biology and management of non‐small cell lung cancer. Nature. 2018;553:446‐454. - PubMed
-
- Du L, Morgensztern D. Chemotherapy for advanced‐stage non–small cell lung cancer. Cancer J. 2015;21:366‐370. - PubMed
-
- Fennell D, Summers Y, Cadranel J, et al. Cisplatin in the modern era: the backbone of first‐line chemotherapy for non‐small cell lung cancer. Cancer Treat Rev. 2016;44:42‐50. - PubMed
-
- Sève P, Dumontet C. Chemoresistance in non‐small cell lung cancer. Curr Med Chem Anti‐Cancer Agents. 2005;5:73‐88. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

