The estimation of additive genetic variance of body size in a wild passerine is sensitive to the method used to estimate relatedness among the individuals
- PMID: 38352200
- PMCID: PMC10862163
- DOI: 10.1002/ece3.10981
The estimation of additive genetic variance of body size in a wild passerine is sensitive to the method used to estimate relatedness among the individuals
Abstract
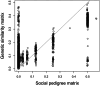
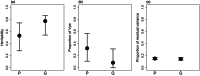
Assessing additive genetic variance is a crucial step in predicting the evolutionary response of a target trait. However, the estimated genetic variance may be sensitive to the methodology used, e.g., the way relatedness is assessed among the individuals, especially in wild populations where social pedigrees can be inaccurate. To investigate this possibility, we investigated the additive genetic variance in tarsus length, a major proxy of skeletal body size in birds. The model species was the collared flycatcher (Ficedula albicollis), a socially monogamous but genetically polygamous migratory passerine. We used two relatedness matrices to estimate the genetic variance: (1) based solely on social links and (2) a genetic similarity matrix based on a large array of single-nucleotide polymorphisms (SNPs). Depending on the relatedness matrix considered, we found moderate to high additive genetic variance and heritability estimates for tarsus length. In particular, the heritability estimates were higher when obtained with the genetic similarity matrix instead of the social pedigree. Our results confirm the potential for this crucial trait to respond to selection and highlight methodological concerns when calculating additive genetic variance and heritability in phenotypic traits. We conclude that using a social pedigree instead of a genetic similarity matrix to estimate relatedness among individuals in a genetically polygamous wild population may significantly deflate the estimates of additive genetic variation.
Keywords: animal model; bird; evolution; quantitative genetics.
© 2024 The Authors. Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Heritability estimates from genomewide relatedness matrices in wild populations: Application to a passerine, using a small sample size.Mol Ecol Resour. 2018 Jul;18(4):838-853. doi: 10.1111/1755-0998.12886. Epub 2018 May 4. Mol Ecol Resour. 2018. PMID: 29667373
-
Moderate heritability and low evolvability of sperm morphology in a species with high risk of sperm competition, the collared flycatcher Ficedula albicollis.J Evol Biol. 2019 Mar;32(3):205-217. doi: 10.1111/jeb.13404. Epub 2018 Dec 10. J Evol Biol. 2019. PMID: 30449037
-
Low genetic variance in the duration of the incubation period in a collared flycatcher (Ficedula albicollis) population.Am Nat. 2012 Jan;179(1):132-6. doi: 10.1086/663193. Epub 2011 Nov 30. Am Nat. 2012. PMID: 22173466
-
RAD-sequencing for estimating genomic relatedness matrix-based heritability in the wild: A case study in roe deer.Mol Ecol Resour. 2019 Sep;19(5):1205-1217. doi: 10.1111/1755-0998.13031. Epub 2019 Jun 12. Mol Ecol Resour. 2019. PMID: 31058463
-
A meta-analysis on the heritability of vertebrate telomere length.J Evol Biol. 2022 Oct;35(10):1283-1295. doi: 10.1111/jeb.14071. Epub 2022 Aug 6. J Evol Biol. 2022. PMID: 35932478 Free PMC article. Review.
Cited by
-
Comparison of Six Measures of Genetic Similarity of Interspecific Brassicaceae Hybrids F2 Generation and Their Parental Forms Estimated on the Basis of ISSR Markers.Genes (Basel). 2024 Aug 23;15(9):1114. doi: 10.3390/genes15091114. Genes (Basel). 2024. PMID: 39336706 Free PMC article.
References
-
- 2019. “Picard toolkit”, in: Broad Institute, GitHub repository. Broad Institute.
-
- Åkesson, M. , Bensch, S. , Hasselquist, D. , Tarka, M. , & Hansson, B. (2008). Estimating heritabilities and genetic correlations: Comparing the ‘animal model’ with parent‐offspring regression using data from a natural population. PLoS One, 3(3), e1739. 10.1371/journal.pone.0001739 - DOI - PMC - PubMed
-
- Alatalo, R. V. , Gustafsson, L. , & Lundberg, A. (1984). High frequency of cuckoldry in pied and collared flycatchers. Oikos, 42(1), 41–47. 10.2307/3544607 - DOI
-
- Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Associated data
LinkOut - more resources
Full Text Sources

