This is a preprint.
Loss of symmetric cell division of apical neural progenitors drives DENND5A-related developmental and epileptic encephalopathy
- PMID: 38352438
- PMCID: PMC10863025
- DOI: 10.1101/2022.08.23.22278845
Loss of symmetric cell division of apical neural progenitors drives DENND5A-related developmental and epileptic encephalopathy
Update in
-
Loss of symmetric cell division of apical neural progenitors drives DENND5A-related developmental and epileptic encephalopathy.Nat Commun. 2024 Aug 22;15(1):7239. doi: 10.1038/s41467-024-51310-z. Nat Commun. 2024. PMID: 39174524 Free PMC article.
Abstract
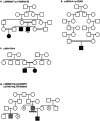
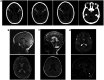
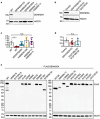
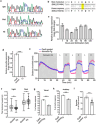
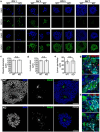
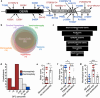
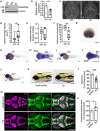
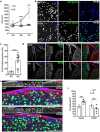
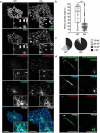
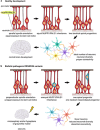
Developmental and epileptic encephalopathies (DEEs) are a heterogenous group of epilepsies in which altered brain development leads to developmental delay and seizures, with the epileptic activity further negatively impacting neurodevelopment. Identifying the underlying cause of DEEs is essential for progress toward precision therapies. Here we describe a group of individuals with biallelic variants in DENND5A and determine that variant type is correlated with disease severity. We demonstrate that DENND5A interacts with MUPP1 and PALS1, components of the Crumbs apical polarity complex, which is required for both neural progenitor cell identity and the ability of these stem cells to divide symmetrically. Induced pluripotent stem cells lacking DENND5A fail to undergo symmetric cell division during neural induction and have an inherent propensity to differentiate into neurons, and transgenic DENND5A mice, with phenotypes like the human syndrome, have an increased number of neurons in the adult subventricular zone. Disruption of symmetric cell division following loss of DENND5A results from misalignment of the mitotic spindle in apical neural progenitors. A subset of DENND5A is localized to centrosomes, which define the spindle poles during mitosis. Cells lacking DENND5A orient away from the proliferative apical domain surrounding the ventricles, biasing daughter cells towards a more fate-committed state and ultimately shortening the period of neurogenesis. This study provides a mechanism behind DENND5A-related DEE that may be generalizable to other developmental conditions and provides variant-specific clinical information for physicians and families.
Conflict of interest statement
Competing Interests KM and RP are employed by GeneDx, LLC. All other authors report no conflicts of interest.
Figures




References
Method References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
