Differential age-related transcriptomic analysis of ovarian granulosa cells in Kazakh horses
- PMID: 38352714
- PMCID: PMC10863452
- DOI: 10.3389/fendo.2024.1346260
Differential age-related transcriptomic analysis of ovarian granulosa cells in Kazakh horses
Abstract
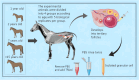
Introduction: The Kazakh horse, renowned for its excellence as a breed, exhibits distinctive reproductive traits characterized by early maturity and seasonal estrus. While normal reproductive function is crucial for ensuring the breeding and expansion of the Kazakh horse population, a noteworthy decline in reproductive capabilities is observed after reaching 14 years of age.
Methods: In this study, ovarian granulosa cells (GCs) were meticulously collected from Kazakh horses aged 1, 2, 7, and above 15 years old (excluding 15 years old) for whole transcriptome sequencing.
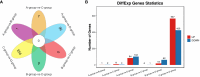
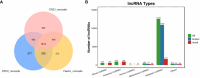
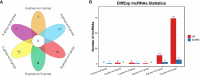
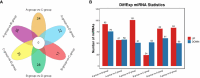
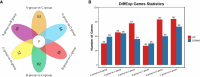
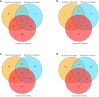
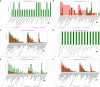
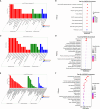
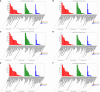
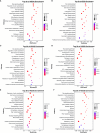
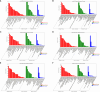
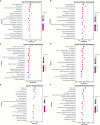
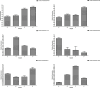
Results: The analysis identified and selected differentially expressed mRNAs, lncRNAs, miRNAs, and circRNAs for each age group, followed by a thorough examination through GO enrichment analysis. The study uncovered significant variations in the expression profiles of mRNAs, lncRNAs, miRNAs, and circRNAs within GCs at different stages of maturity. Notably, eca-miR-486-3p and miR-486-y exhibited the highest degree of connectivity. Subsequent GO, KEGG, PPI, and ceRNA network analyses elucidated that the differentially expressed target genes actively participate in signaling pathways associated with cell proliferation, apoptosis, and hormonal regulation. These pathways include but are not limited to the MAPK signaling pathway, Hippo signaling pathway, Wnt signaling pathway, Calcium signaling pathway, Aldosterone synthesis and secretion, Cellular senescence, and NF-kappa B signaling pathway-essentially encompassing signal transduction pathways crucial to reproductive processes.
Discussion: This research significantly contributes to unraveling the molecular mechanisms governing follicular development in Kazakh horses. It establishes and preliminarily validates a differential regulatory network involving lncRNA-miRNA-mRNA, intricately associated with processes such as cell proliferation, differentiation, and apoptosis and integral to the developmental intricacies of stromal follicles. The findings of this study provide a solid theoretical foundation for delving deeper into the realm of reproductive aging in Kazakh mares, presenting itself as a pivotal regulatory pathway in the context of horse ovarian development.
Keywords: PPI; ceRNA; horse; ovarian granulosa cell; whole transcriptome.
Copyright © 2024 Ren, Wang, Zeng, Wang, Meng and Yao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
mRNA, lncRNA and Circular RNA Expression Profiles in Granulosa Cells of Infertile Women with Ovarian Endometriosis.Reprod Sci. 2022 Oct;29(10):2937-2946. doi: 10.1007/s43032-022-00966-3. Epub 2022 Jul 7. Reprod Sci. 2022. PMID: 35799021
-
Preliminary construction of non-coding RNAs and ceRNA regulatory networks mediated by exosomes in porcine follicular fluid.Genomics. 2024 Sep;116(5):110920. doi: 10.1016/j.ygeno.2024.110920. Epub 2024 Aug 14. Genomics. 2024. PMID: 39151553
-
Comprehensive analysis of the whole-transcriptome landscape of the ovarian cortex from Mongolian horses that reproduce seasonally.Comp Biochem Physiol Part D Genomics Proteomics. 2024 Mar;49:101179. doi: 10.1016/j.cbd.2023.101179. Epub 2023 Dec 15. Comp Biochem Physiol Part D Genomics Proteomics. 2024. PMID: 38134534
-
Differential analysis of testicular LncRNA in Kazakh horses of different ages.Int J Biol Macromol. 2025 Sep;321(Pt 1):146228. doi: 10.1016/j.ijbiomac.2025.146228. Epub 2025 Jul 22. Int J Biol Macromol. 2025. PMID: 40706934
-
Whole transcriptome analysis and construction of gene regulatory networks of granulosa cells from patients with polycystic ovary syndrome (PCOS).Eur J Med Res. 2025 Jan 7;30(1):9. doi: 10.1186/s40001-024-02237-0. Eur J Med Res. 2025. PMID: 39773546 Free PMC article.
Cited by
-
Transcriptome and chromatin accessibility landscape of ovarian development at different egg-laying stages in taihe black-bone silky fowls.Poult Sci. 2025 Mar;104(3):104864. doi: 10.1016/j.psj.2025.104864. Epub 2025 Feb 7. Poult Sci. 2025. PMID: 39922133 Free PMC article.
-
Transcriptomic sequencing and differential analysis of Kazakh horse muscles from various anatomical locations.Front Vet Sci. 2025 Jul 24;12:1633786. doi: 10.3389/fvets.2025.1633786. eCollection 2025. Front Vet Sci. 2025. PMID: 40777832 Free PMC article.
-
Circular RNAs as regulators and biomarkers of mammalian ovarian ageing.Geroscience. 2025 Jul 17. doi: 10.1007/s11357-025-01798-0. Online ahead of print. Geroscience. 2025. PMID: 40676447 Review.
-
Prediction of the molecular action of Trypanosoma vivax on bovine reproductive parameters and risk factors associated with trypanosomiasis in northern Minas Gerais, Brazil.Vet World. 2025 Apr;18(4):837-850. doi: 10.14202/vetworld.2025.837-850. Epub 2025 Apr 19. Vet World. 2025. PMID: 40453953 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

