ChipFilter: Microfluidic-Based Comprehensive Sample Preparation Methodology for Microbial Consortia
- PMID: 38353246
- PMCID: PMC10913871
- DOI: 10.1021/acs.jproteome.3c00288
ChipFilter: Microfluidic-Based Comprehensive Sample Preparation Methodology for Microbial Consortia
Abstract
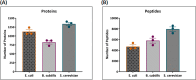
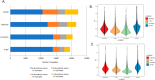
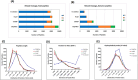
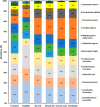
The metaproteomic approach is an attractive way to describe a microbiome at the functional level, allowing the identification and quantification of proteins across a broad dynamic range as well as the detection of post-translational modifications. However, it remains relatively underutilized, mainly due to technical challenges that should be addressed, including the complexity of extracting proteins from heterogeneous microbial communities. Here, we show that a ChipFilter microfluidic device coupled to a liquid chromatography tandem mass spectrometry (LC-MS/MS) setup can be successfully used for the identification of microbial proteins. Using cultures of Escherichia coli, Bacillus subtilis, and Saccharomyces cerevisiae, we have shown that it is possible to directly lyse the cells and digest the proteins in the ChipFilter to allow the identification of a higher number of proteins and peptides than that by standard protocols, even at low cell density. The peptides produced are overall longer after ChipFilter digestion but show no change in their degree of hydrophobicity. Analysis of a more complex mixture of 17 species from the gut microbiome showed that the ChipFilter preparation was able to identify and estimate the amounts of 16 of these species. These results show that ChipFilter can be used for the proteomic study of microbiomes, particularly in the case of a low volume or cell density. The mass spectrometry data have been deposited on the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD039581.
Keywords: metaproteomic; proteomic; sample preparation.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



Similar articles
-
A single microfluidic device for multi-omics analysis sample preparation.Lab Chip. 2025 Feb 11;25(4):590-599. doi: 10.1039/d4lc00919c. Lab Chip. 2025. PMID: 39820672
-
On-Chip Sample Preparation Using a ChipFilter Coupled to NanoLC-MS/MS for Bottom-Up Proteomics.J Proteome Res. 2020 Jul 2;19(7):2654-2663. doi: 10.1021/acs.jproteome.9b00832. Epub 2020 May 8. J Proteome Res. 2020. PMID: 32343577
-
Automation of peptide desalting for proteomic liquid chromatography - tandem mass spectrometry by centrifugal microfluidics.Lab Chip. 2021 Jun 1;21(11):2255-2264. doi: 10.1039/d1lc00137j. Lab Chip. 2021. PMID: 33908535
-
[Advances in high-throughput proteomic analysis].Se Pu. 2021 Feb;39(2):112-117. doi: 10.3724/SP.J.1123.2020.08023. Se Pu. 2021. PMID: 34227342 Free PMC article. Review. Chinese.
-
[Recent progress in capillary electrophoresis-based high-sensitivity proteomics].Se Pu. 2020 Oct 8;38(10):1125-1132. doi: 10.3724/SP.J.1123.2020.03003. Se Pu. 2020. PMID: 34213109 Review. Chinese.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

