Beyond the tail: the consequence of context in histone post-translational modification and chromatin research
- PMID: 38353483
- PMCID: PMC10903488
- DOI: 10.1042/BCJ20230342
Beyond the tail: the consequence of context in histone post-translational modification and chromatin research
Abstract
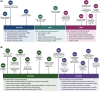
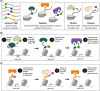
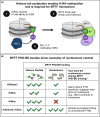
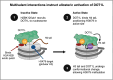
The role of histone post-translational modifications (PTMs) in chromatin structure and genome function has been the subject of intense debate for more than 60 years. Though complex, the discourse can be summarized in two distinct - and deceptively simple - questions: What is the function of histone PTMs? And how should they be studied? Decades of research show these queries are intricately linked and far from straightforward. Here we provide a historical perspective, highlighting how the arrival of new technologies shaped discovery and insight. Despite their limitations, the tools available at each period had a profound impact on chromatin research, and provided essential clues that advanced our understanding of histone PTM function. Finally, we discuss recent advances in the application of defined nucleosome substrates, the study of multivalent chromatin interactions, and new technologies driving the next era of histone PTM research.
Keywords: chromatin; histone code; histone peptides; histone post-translational modifications; histones; nucleosome.
© 2024 The Author(s).
Conflict of interest statement
EpiCypher is a commercial developer and supplier of reagents and technologies used for chromatin studies. The authors are employed by (and own shares in) EpiCypher. M.-C.K. is a board member of EpiCypher.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

