Genetic epidemiology and plasmid-mediated transmission of mcr-1 by Escherichia coli ST155 from wastewater of long-term care facilities
- PMID: 38353552
- PMCID: PMC10913736
- DOI: 10.1128/spectrum.03707-23
Genetic epidemiology and plasmid-mediated transmission of mcr-1 by Escherichia coli ST155 from wastewater of long-term care facilities
Abstract
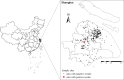
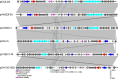
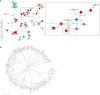
Long-term care facilities (LTCFs) for older people play an important and unique role in multidrug-resistant organism transmission. Herein, we investigated the genetic characteristics of mobile colistin resistance gene (mcr-1)-carrying Escherichia coli strains isolated from wastewater of LTCFs in Shanghai. Antimicrobial susceptibility test was carried out by agar dilution methods. Whole-genome sequencing and plasmid sequencing were conducted, and resistance genes and sequence types of colistin in E. coli isolates were analyzed. Core genome multilocus sequence typing (cgMLST) analysis was performed by the Ridom SeqSphere+ software. Phylogenetic tree through the maximum likelihood method was constructed by MEGA X. Out of 306 isolates, only 1 E. coli named ECSJ33 was found, and the plasmid pECSJ33 from ECSJ33 harbored the mcr-1 gene that was located with 59,080 bp belonging to IncI2 type. The plasmid pECSJ33 was capable of conjugation with an efficiency of 2.9 × 10-2. Bioinformatic analysis indicated pECSJ33 shared backbone with the previously reported mcr-1-harboring pHNGDF93 isolated from fish source. Moreover, the cgMLST analysis revealed that ECSJ33 belongs to different lineages from those reported from previous E. coli strains but shared high similarity to NCTC11129 in cluster 11. The phylogenetic tree revealed MCR-1 of ECSJ33 in this study was mostly of animal food origin and that they were closely related. Our study firstly reports detection of genome sequence of a multidrug-resistant mcr-1-harboring E. coli ST155 from wastewater of LTCF source in China. The data may prove that the plasmid pECSJ33 belongs to food origin and help to understand the antimicrobial resistance mechanisms and genomic features of colistin resistance under One Health approach.IMPORTANCEOne Escherichia coli named ECSJ33 was found from wastewater of a long-term care facility (LTCF) and the plasmid pECSJ33 from ECSJ33 harbored the mobile colistin resistance gene (mcr-1) that was located with 59,080 bp belonging to IncI2 type, which was capable of conjugation with an efficiency of 2.9 × 10-2. This paper firstly reports an mcr-1-carrying E. coli strain ST155 isolated from LTCF in China. Comparative genomics analysis indicated pECSJ33 shared backbone with the previously reported mcr-1-harboring pHNGDF93 isolated from fish source. The phylogenetic tree revealed MCR-1 protein of ECSJ33 in this study was mostly of animal food origin and that they were closely related. Therefore, the pECSJ33 could be considered as food-origin transmission mcr-1-harboring plasmid.
Keywords: Escherichia coli ST155; long-term-care facility; mcr-1; plasmid; wastewater.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7 - DOI - PubMed
-
- Shen Y, Zhou H, Xu J, Wang Y, Zhang Q, Walsh TR, Shao B, Wu C, Hu Y, Yang L, Shen Z, Wu Z, Sun Q, Ou Y, Wang Y, Wang S, Wu Y, Cai C, Li J, Shen J, Zhang R, Wang Y. 2018. Anthropogenic and environmental factors associated with high incidence of mcr-1 carriage in humans across China. Nat Microbiol 3:1054–1062. doi: 10.1038/s41564-018-0205-8 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

