Lsr2, a pleiotropic regulator at the core of the infectious strategy of Mycobacterium abscessus
- PMID: 38353553
- PMCID: PMC10913753
- DOI: 10.1128/spectrum.03528-23
Lsr2, a pleiotropic regulator at the core of the infectious strategy of Mycobacterium abscessus
Abstract
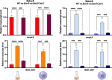
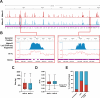
Mycobacterium abscessus is a non-tuberculous mycobacterium, causing lung infections in cystic fibrosis patients. During pulmonary infection, M. abscessus switches from smooth (Mabs-S) to rough (Mabs-R) morphotypes, the latter being hyper-virulent. Previously, we isolated the lsr2 gene as differentially expressed during S-to-R transition. lsr2 encodes a pleiotropic transcription factor that falls under the superfamily of nucleoid-associated proteins. Here, we used two functional genomic methods, RNA-seq and chromatin immunoprecipitation-sequencing (ChIP-seq), to elucidate the molecular role of Lsr2 in the pathobiology of M. abscessus. Transcriptomic analysis shows that Lsr2 differentially regulates gene expression across both morphotypes, most of which are involved in several key cellular processes of M. abscessus, including host adaptation and antibiotic resistance. These results were confirmed through quantitative real-time PCR, as well as by minimum inhibitory concentration tests and infection tests on macrophages in the presence of antibiotics. ChIP-seq analysis revealed that Lsr2 extensively binds the M. abscessus genome at AT-rich sequences and appears to form long domains that participate in the repression of its target genes. Unexpectedly, the genomic distribution of Lsr2 revealed no distinctions between Mabs-S and Mabs-R, implying more intricate mechanisms at play for achieving target selectivity.IMPORTANCELsr2 is a crucial transcription factor and chromosome organizer involved in intracellular growth and virulence in the smooth and rough morphotypes of Mycobacterium abscessus. Using RNA-seq and chromatin immunoprecipitation-sequencing (ChIP-seq), we investigated the molecular role of Lsr2 in gene expression regulation along with its distribution on M. abscessus genome. Our study demonstrates the pleiotropic regulatory role of Lsr2, regulating the expression of many genes coordinating essential cellular and molecular processes in both morphotypes. In addition, we have elucidated the role of Lsr2 in antibiotic resistance both in vitro and in vivo, where lsr2 mutant strains display heightened sensitivity to antibiotics. Through ChIP-seq, we reported the widespread distribution of Lsr2 on M. abscessus genome, revealing a direct repressive effect due to its extensive binding on promoters or coding sequences of its targets. This study unveils the significant regulatory role of Lsr2, intricately intertwined with its function in shaping the organization of the M. abscessus genome.
Keywords: Lsr2; Mycobacterium abscessus; S/R morphotypes; antibiotic resistance; nucleoid-associated protein; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Qvist T, Taylor-Robinson D, Waldmann E, Olesen HV, Hansen CR, Mathiesen IH, Høiby N, Katzenstein TL, Smyth RL, Diggle PJ, Pressler T. 2016. Comparing the harmful effects of nontuberculous mycobacteria and Gram negative bacteria on lung function in patients with cystic fibrosis. J Cyst Fibros 15:380–385. doi: 10.1016/j.jcf.2015.09.007 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

