Generation of Human Isogenic Induced Pluripotent Stem Cell Lines with CRISPR Prime Editing
- PMID: 38353623
- PMCID: PMC10880268
- DOI: 10.1089/crispr.2023.0066
Generation of Human Isogenic Induced Pluripotent Stem Cell Lines with CRISPR Prime Editing
Abstract
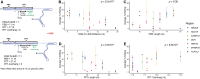
We developed an efficient CRISPR prime editing protocol and generated isogenic-induced pluripotent stem cell (iPSC) lines carrying heterozygous or homozygous alleles for putatively causal single nucleotide variants at six type 2 diabetes loci (ABCC8, MTNR1B, TCF7L2, HNF4A, CAMK1D, and GCK). Our two-step sequence-based approach to first identify transfected cell pools with the highest fraction of edited cells significantly reduced the downstream efforts to isolate single clones of edited cells. We found that prime editing can make targeted genetic changes in iPSC and optimization of system components and guide RNA designs that were critical to achieve acceptable efficiency. Systems utilizing PEmax, epegRNA modifications, and MLH1dn provided significant benefit, producing editing efficiencies of 36-73%. Editing success and pegRNA design optimization required for each variant differed depending on the sequence at the target site. With attention to design, prime editing is a promising approach to generate isogenic iPSC lines, enabling the study of specific genetic changes in a common genetic background.
Conflict of interest statement
No competing financial interests exist.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

