Type I interferon pathway genetic variants in severe COVID-19
- PMID: 38354910
- PMCID: PMC10901847
- DOI: 10.1016/j.virusres.2024.199339
Type I interferon pathway genetic variants in severe COVID-19
Abstract
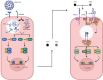
Coronavirus Disease 2019 (COVID-19) is an infectious disease caused by SARS-CoV-2. According to the World Health Organization (WHO), there have been over 760 million reported cases and over 6 million deaths caused by this disease worldwide. The severity of COVID-19 is based on symptoms presented by the patient and is divided as asymptomatic, mild, moderate, severe, and critical. The manifestations are interconnected with genetic variations. The innate immunity is the quickest response mechanism of an organism against viruses. Type I interferon pathway plays a key role in antiviral responses due to viral replication inhibition in infected cells and adaptive immunity stimulation induced by interferon molecules. Thus, variants in type I interferon pathway's genes are being studied in different COVID-19 manifestations. This review summarizes the role of variants in type I interferon pathway's genes on prognosis and severity progression of COVID-19.
Keywords: COVID-19; Single nucleotide polymorphisms; Type I interferon.
Copyright © 2024. Published by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Similar articles
-
Interferon at the crossroads of SARS-CoV-2 infection and COVID-19 disease.J Biol Chem. 2023 Aug;299(8):104960. doi: 10.1016/j.jbc.2023.104960. Epub 2023 Jun 24. J Biol Chem. 2023. PMID: 37364688 Free PMC article. Review.
-
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling.Signal Transduct Target Ther. 2020 Dec 28;5(1):299. doi: 10.1038/s41392-020-00438-7. Signal Transduct Target Ther. 2020. PMID: 33372174 Free PMC article.
-
SARS-CoV-2-Encoded MiRNAs Inhibit Host Type I Interferon Pathway and Mediate Allelic Differential Expression of Susceptible Gene.Front Immunol. 2021 Dec 23;12:767726. doi: 10.3389/fimmu.2021.767726. eCollection 2021. Front Immunol. 2021. PMID: 35003084 Free PMC article.
-
Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2.mBio. 2020 Sep 10;11(5):e01928-20. doi: 10.1128/mBio.01928-20. mBio. 2020. PMID: 32913009 Free PMC article.
-
Type I and III interferon responses in SARS-CoV-2 infection.Exp Mol Med. 2021 May;53(5):750-760. doi: 10.1038/s12276-021-00592-0. Epub 2021 May 6. Exp Mol Med. 2021. PMID: 33953323 Free PMC article. Review.
Cited by
-
Genetic Predictors of Paxlovid Treatment Response: The Role of IFNAR2, OAS1, OAS3, and ACE2 in COVID-19 Clinical Course.J Pers Med. 2025 Apr 17;15(4):156. doi: 10.3390/jpm15040156. J Pers Med. 2025. PMID: 40278335 Free PMC article.
-
Association of IFNAR2 and TYK2 with COVID-19 pathology: current and future.Front Immunol. 2024 Sep 16;15:1462628. doi: 10.3389/fimmu.2024.1462628. eCollection 2024. Front Immunol. 2024. PMID: 39351231 Free PMC article. No abstract available.
-
Deciphering TLR and JAK/STAT pathways: genetic variants and targeted therapies in COVID-19.Mol Biol Rep. 2025 Jul 18;52(1):733. doi: 10.1007/s11033-025-10843-2. Mol Biol Rep. 2025. PMID: 40679664 Review.
-
SARS-CoV-2 Impairs Osteoblast Differentiation Through Spike Glycoprotein and Cytokine Dysregulation.Viruses. 2025 Jan 22;17(2):143. doi: 10.3390/v17020143. Viruses. 2025. PMID: 40006897 Free PMC article.
-
MDA5 Is a Major Determinant of Developing Symptoms in Critically Ill COVID-19 Patients.Clin Rev Allergy Immunol. 2024 Dec;67(1-3):58-72. doi: 10.1007/s12016-024-09008-z. Epub 2024 Oct 26. Clin Rev Allergy Immunol. 2024. PMID: 39460899 Review.
References
-
- Abdelhafez M., Nasereddin A., Shamma O.A., Abed R., Sinnokrot R., Marof O., Heif T., Erekat Z., Al-Jawabreh A., Ereqat S. Association of IFNAR2 rs2236757 and OAS3 rs10735079 polymorphisms with susceptibility to COVID-19 infection and severity in palestine. Interdiscip. Perspect. Infect. Dis. 2023;11 doi: 10.1155/2023/9551163. 2023. - DOI - PMC - PubMed
-
- Abolhassani H., Landegren N., Bastard P., Materna M., Modaresi M., Du L., Aranda-Guillén M., Sardh F., Zuo F., Zhang P., Marcotte H., Marr N., Khan T., Ata M., Al-Ali F., Pescarmona R., Belot A., Béziat V., Zhang Q., Casanova J.L., Kämpe O., Zhang S.Y., Hammarström L., Pan-Hammarström Q. Inherited IFNAR1 deficiency in a child with both critical COVID-19 pneumonia and multisystem inflammatory syndrome. J. Clin. Immunol. 2022;42:471–483. doi: 10.1007/s10875-022-01215-7. - DOI - PMC - PubMed
-
- Azkur A.K., Akdis M., Azkur D., Sokolowska M., van de Veen W., Brüggen M.C., O'Mahony L., Gao Y., Nadeau K., Akdis C.A. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy Eur. J. Allergy Clin. Immunol. 2020;75:1564–1581. doi: 10.1111/all.14364. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

