Efficient derivation of embryonic stem cells and primordial germ cell-like cells in cattle
- PMID: 38355134
- PMCID: PMC11017101
- DOI: 10.1262/jrd.2023-087
Efficient derivation of embryonic stem cells and primordial germ cell-like cells in cattle
Abstract
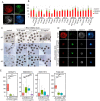
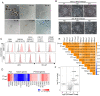
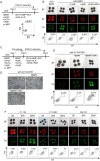
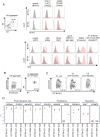
The induction of the germ cell lineage from pluripotent stem cells (in vitro gametogenesis) will help understand the mechanisms underlying germ cell differentiation and provide an alternative source of gametes for reproduction. This technology is especially important for cattle, which are among the most important livestock species for milk and meat production. Here, we developed a new method for robust induction of primordial germ cell-like cells (PGCLCs) from newly established bovine embryonic stem (bES) cells. First, we refined the pluripotent culture conditions for pre-implantation embryos and ES cells. Inhibition of RHO increased the number of epiblast cells in the pre-implantation embryos and dramatically improved the efficiency of ES cell establishment. We then determined suitable culture conditions for PGCLC differentiation using bES cells harboring BLIMP1-tdTomato and TFAP2C-mNeonGreen (BTTN) reporter constructs. After a 24-h culture with bone morphogenetic protein 4 (BMP4), followed by three-dimensional culture with BMP4 and a chemical agonist and WNT signaling chemical antagonist, bES cells became positive for the reporters. A set of primordial germ cells (PGC) marker genes, including PRDM1/BLIMP1, TFAP2C, SOX17, and NANOS3, were expressed in BTTN-positive cells. These bovine PGCLCs (bPGCLCs) were isolated as KIT/CD117-positive and CD44-negative cell populations. We anticipate that this method for the efficient establishment of bES cells and induction of PGCLCs will be useful for stem cell-based reproductive technologies in cattle.
Keywords: Blastocyst; Pluripotent stem cells; Primordial germ cells.
Conflict of interest statement
We declare that there are no conflicts of interest that could be perceived as prejudicing the impartiality of the reported research.
Figures

References
-
- Saitou M, Hayashi K. Mammalian in vitro gametogenesis. Science 2021; 374: eaaz6830. - PubMed
-
- Oikawa M, Kobayashi H, Sanbo M, Mizuno N, Iwatsuki K, Takashima T, Yamauchi K, Yoshida F, Yamamoto T, Shinohara T, Nakauchi H, Kurimoto K, Hirabayashi M, Kobayashi T. Functional primordial germ cell-like cells from pluripotent stem cells in rats. Science 2022; 376: 176–179. - PubMed
-
- Kobayashi T, Castillo-Venzor A, Penfold CA, Morgan M, Mizuno N, Tang WWC, Osada Y, Hirao M, Yoshida F, Sato H, Nakauchi H, Hirabayashi M, Surani MA. Tracing the emergence of primordial germ cells from bilaminar disc rabbit embryos and pluripotent stem cells. Cell Rep 2021; 37: 109812. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

