The IL6/JAK/STAT3 signaling axis is a therapeutic vulnerability in SMARCB1-deficient bladder cancer
- PMID: 38355560
- PMCID: PMC10867091
- DOI: 10.1038/s41467-024-45132-2
The IL6/JAK/STAT3 signaling axis is a therapeutic vulnerability in SMARCB1-deficient bladder cancer
Abstract
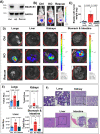
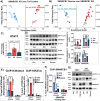
SMARCB1 loss has long been observed in many solid tumors. However, there is a need to elucidate targetable pathways driving growth and metastasis in SMARCB1-deficient tumors. Here, we demonstrate that SMARCB1 deficiency, defined as genomic SMARCB1 copy number loss associated with reduced mRNA, drives disease progression in patients with bladder cancer by engaging STAT3. SMARCB1 loss increases the chromatin accessibility of the STAT3 locus in vitro. Orthotopically implanted SMARCB1 knockout (KO) cell lines exhibit increased tumor growth and metastasis. SMARCB1-deficient tumors show an increased IL6/JAK/STAT3 signaling axis in in vivo models and patients. Furthermore, a pSTAT3 selective inhibitor, TTI-101, reduces tumor growth in SMARCB1 KO orthotopic cell line-derived xenografts and a SMARCB1-deficient patient derived xenograft model. We have identified a gene signature generated from SMARCB1 KO tumors that predicts SMARCB1 deficiency in patients. Overall, these findings support the clinical evaluation of STAT3 inhibitors for the treatment of SMARCB1-deficient bladder cancer.
© 2024. The Author(s).
Conflict of interest statement
S.M.K. is stakeholder of NeoZenome Therapeutics Inc. P.M. has received honoraria for service on a Scientific Advisory Board for Mirati Therapeutics, Bristol Myers Squibb, and Exelixis; consulting for Axiom Healthcare Strategies; non-branded educational programs supported by Exelixis and Pfizer; and research funding for clinical trials from Takeda, Bristol Myers Squibb, Mirati Therapeutics, Gateway for Cancer Research, and The University of Texas MD Anderson Cancer Center. M.J.E. reports grants from CPRIT and McNair Medical Foundation during the conduct of the study; personal fees from AstraZeneca outside the submitted work; in addition, M.J.E. has a patent for PAM50 issued, licensed, and with royalties paid from Veracyte. M.J.E. is current employee of AstraZeneca. H.C.H. is a consultant for Avenge Bio, Inc. A.S.K. is Scientific Advisor to Karkinos Health Care Pvt Ltd, India and unpaid visiting faculty to Sri Sathya Sai Institute for Higher Learning, India. The other authors declare that they have no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

